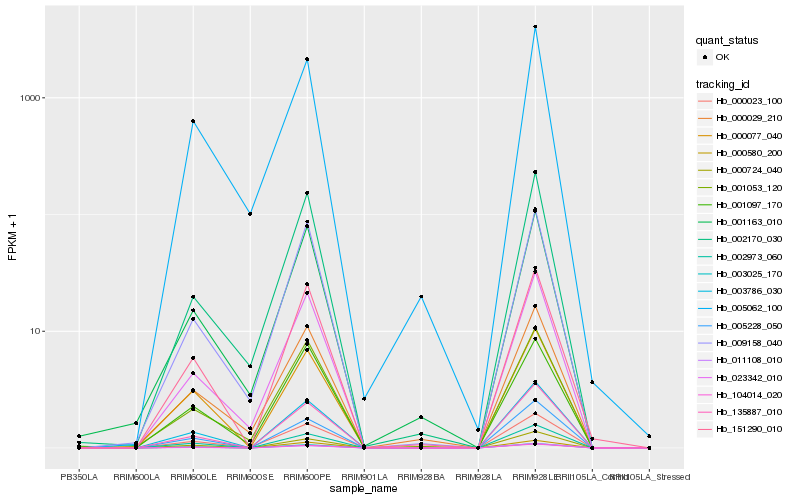
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000023_100 |
0.0 |
- |
- |
Mechanosensitive channel of small conductance-like 10 [Theobroma cacao] |
| 2 |
Hb_023342_010 |
0.0381202539 |
- |
- |
- |
| 3 |
Hb_003786_030 |
0.0538342106 |
- |
- |
BnaUnng00830D [Brassica napus] |
| 4 |
Hb_001053_120 |
0.0612069551 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46-like [Populus euphratica] |
| 5 |
Hb_002973_060 |
0.0624598421 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_009158_040 |
0.0634752045 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |
| 7 |
Hb_135887_010 |
0.0637844078 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 8 |
Hb_000029_210 |
0.0656934267 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 9 |
Hb_000724_040 |
0.065722162 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein CMB1-like [Jatropha curcas] |
| 10 |
Hb_104014_020 |
0.0668221675 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39020 isoform X2 [Jatropha curcas] |
| 11 |
Hb_002170_030 |
0.0679905283 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 12 |
Hb_005228_050 |
0.0694688374 |
- |
- |
PREDICTED: uncharacterized protein LOC104896168 [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_005062_100 |
0.0704263276 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_151290_010 |
0.0732238269 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_000580_200 |
0.0737408243 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003025_170 |
0.0739597995 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g59900 [Jatropha curcas] |
| 17 |
Hb_001097_170 |
0.0763378519 |
- |
- |
PREDICTED: protein NDR1 [Jatropha curcas] |
| 18 |
Hb_011108_010 |
0.0764932259 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103335582 [Prunus mume] |
| 19 |
Hb_001163_010 |
0.0769934448 |
- |
- |
PREDICTED: syncoilin [Jatropha curcas] |
| 20 |
Hb_000077_040 |
0.0803423277 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |