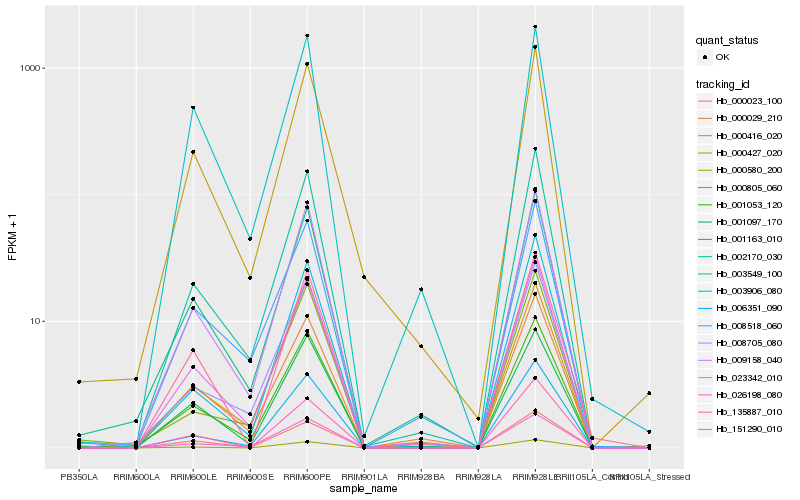
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009158_040 |
0.0 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |
| 2 |
Hb_001053_120 |
0.0238698369 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46-like [Populus euphratica] |
| 3 |
Hb_002170_030 |
0.0446624301 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 4 |
Hb_026198_080 |
0.0454997001 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 [Populus euphratica] |
| 5 |
Hb_023342_010 |
0.0460850625 |
- |
- |
- |
| 6 |
Hb_006351_090 |
0.0565723644 |
- |
- |
UDP-glycosyltransferase 85A1 [Gossypium arboreum] |
| 7 |
Hb_001163_010 |
0.0598166596 |
- |
- |
PREDICTED: syncoilin [Jatropha curcas] |
| 8 |
Hb_008705_080 |
0.0621424137 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 9 |
Hb_000023_100 |
0.0634752045 |
- |
- |
Mechanosensitive channel of small conductance-like 10 [Theobroma cacao] |
| 10 |
Hb_000580_200 |
0.0649060396 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001097_170 |
0.0660000372 |
- |
- |
PREDICTED: protein NDR1 [Jatropha curcas] |
| 12 |
Hb_000029_210 |
0.0746679214 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_008518_060 |
0.0795106113 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 1 [Jatropha curcas] |
| 14 |
Hb_000416_020 |
0.0803317974 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000427_020 |
0.0807858194 |
- |
- |
PsbA, partial [Dianthus pygmaeus] |
| 16 |
Hb_151290_010 |
0.0846596038 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_135887_010 |
0.0913390457 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 18 |
Hb_003906_080 |
0.0927633804 |
- |
- |
Peroxidase 66 precursor, putative [Ricinus communis] |
| 19 |
Hb_003549_100 |
0.0940859129 |
- |
- |
PREDICTED: uncharacterized protein LOC105647410 [Jatropha curcas] |
| 20 |
Hb_000805_060 |
0.0943658488 |
- |
- |
laccase, putative [Ricinus communis] |