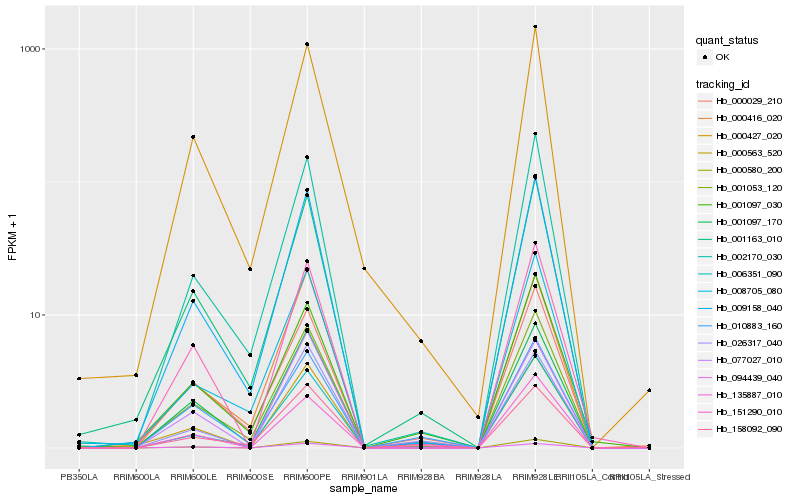
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000563_520 |
0.0 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 2 |
Hb_026317_040 |
0.0889739359 |
- |
- |
hypothetical protein POPTR_0009s12270g [Populus trichocarpa] |
| 3 |
Hb_001163_010 |
0.0890835327 |
- |
- |
PREDICTED: syncoilin [Jatropha curcas] |
| 4 |
Hb_000580_200 |
0.0915191317 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001053_120 |
0.092007063 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46-like [Populus euphratica] |
| 6 |
Hb_009158_040 |
0.0977736289 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |
| 7 |
Hb_006351_090 |
0.0988555346 |
- |
- |
UDP-glycosyltransferase 85A1 [Gossypium arboreum] |
| 8 |
Hb_151290_010 |
0.0996384756 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_001097_030 |
0.108311482 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 10 |
Hb_000416_020 |
0.1087152608 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_010883_160 |
0.1113699439 |
- |
- |
PREDICTED: pathogenesis-related protein 5 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001097_170 |
0.113030929 |
- |
- |
PREDICTED: protein NDR1 [Jatropha curcas] |
| 13 |
Hb_000029_210 |
0.1142289199 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_158092_090 |
0.1156479136 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_002170_030 |
0.1162028996 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 16 |
Hb_077027_010 |
0.1163810649 |
- |
- |
hypothetical protein JCGZ_04511 [Jatropha curcas] |
| 17 |
Hb_008705_080 |
0.1165282731 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 18 |
Hb_000427_020 |
0.1190680962 |
- |
- |
PsbA, partial [Dianthus pygmaeus] |
| 19 |
Hb_135887_010 |
0.1191185569 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 20 |
Hb_094439_040 |
0.1192151699 |
- |
- |
PREDICTED: cytochrome P450 93A1-like [Citrus sinensis] |