| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
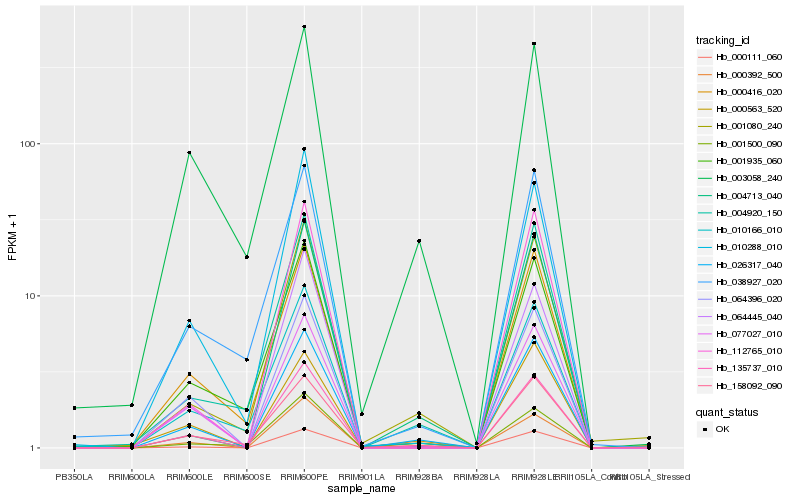
Hb_026317_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0009s12270g [Populus trichocarpa] |
| 2 |
Hb_135737_010 |
0.0856506317 |
- |
- |
hypothetical protein POPTR_0004s02760g [Populus trichocarpa] |
| 3 |
Hb_000111_060 |
0.0884766632 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 6 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000563_520 |
0.0889739359 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 5 |
Hb_001080_240 |
0.0968320903 |
- |
- |
hypothetical protein PHAVU_001G152800g [Phaseolus vulgaris] |
| 6 |
Hb_064396_020 |
0.097490724 |
- |
- |
hypothetical protein POPTR_0019s09950g [Populus trichocarpa] |
| 7 |
Hb_077027_010 |
0.0975312037 |
- |
- |
hypothetical protein JCGZ_04511 [Jatropha curcas] |
| 8 |
Hb_000416_020 |
0.0990823735 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_158092_090 |
0.1002709617 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_004920_150 |
0.1022430056 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_112765_010 |
0.1050609 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004713_040 |
0.1118580197 |
- |
- |
hypothetical protein JCGZ_22567 [Jatropha curcas] |
| 13 |
Hb_001500_090 |
0.1133117931 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_010288_010 |
0.1135770681 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 15 |
Hb_000392_500 |
0.1158777614 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_010166_010 |
0.117583653 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 17 |
Hb_003058_240 |
0.1182591855 |
- |
- |
PREDICTED: tubulin beta-5 chain-like [Jatropha curcas] |
| 18 |
Hb_038927_020 |
0.1190567303 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC13 [Cucumis melo] |
| 19 |
Hb_001935_060 |
0.1198685893 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 20 |
Hb_064445_040 |
0.1209302989 |
- |
- |
hypothetical protein POPTR_0005s09430g [Populus trichocarpa] |