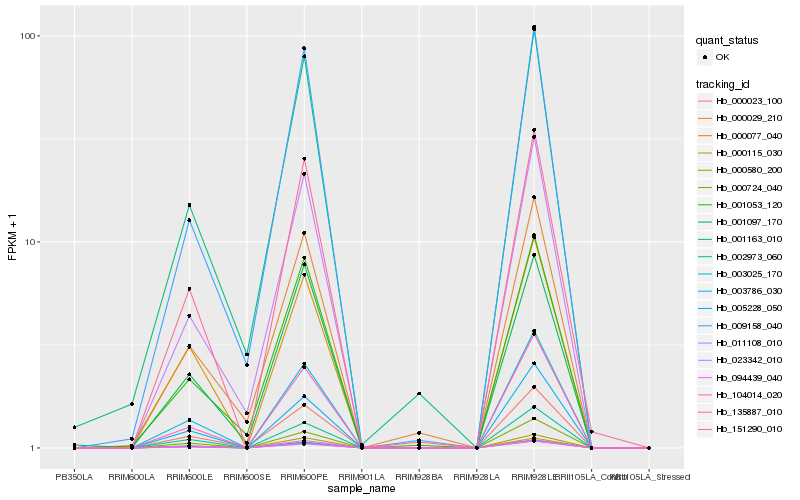
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_151290_010 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_000580_200 |
0.0593412756 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003786_030 |
0.0618256147 |
- |
- |
BnaUnng00830D [Brassica napus] |
| 4 |
Hb_104014_020 |
0.0662853532 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39020 isoform X2 [Jatropha curcas] |
| 5 |
Hb_003025_170 |
0.0671848594 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g59900 [Jatropha curcas] |
| 6 |
Hb_135887_010 |
0.0707612457 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 7 |
Hb_000023_100 |
0.0732238269 |
- |
- |
Mechanosensitive channel of small conductance-like 10 [Theobroma cacao] |
| 8 |
Hb_002973_060 |
0.0735113922 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000724_040 |
0.0808246919 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein CMB1-like [Jatropha curcas] |
| 10 |
Hb_001097_170 |
0.0829405989 |
- |
- |
PREDICTED: protein NDR1 [Jatropha curcas] |
| 11 |
Hb_001053_120 |
0.08427317 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46-like [Populus euphratica] |
| 12 |
Hb_000077_040 |
0.0843701528 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 13 |
Hb_009158_040 |
0.0846596038 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |
| 14 |
Hb_005228_050 |
0.0856216436 |
- |
- |
PREDICTED: uncharacterized protein LOC104896168 [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_023342_010 |
0.0872532219 |
- |
- |
- |
| 16 |
Hb_011108_010 |
0.0905400513 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103335582 [Prunus mume] |
| 17 |
Hb_000115_030 |
0.0958968865 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 42 [Jatropha curcas] |
| 18 |
Hb_094439_040 |
0.0962158156 |
- |
- |
PREDICTED: cytochrome P450 93A1-like [Citrus sinensis] |
| 19 |
Hb_001163_010 |
0.0963627006 |
- |
- |
PREDICTED: syncoilin [Jatropha curcas] |
| 20 |
Hb_000029_210 |
0.0976582863 |
- |
- |
protein binding protein, putative [Ricinus communis] |