| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
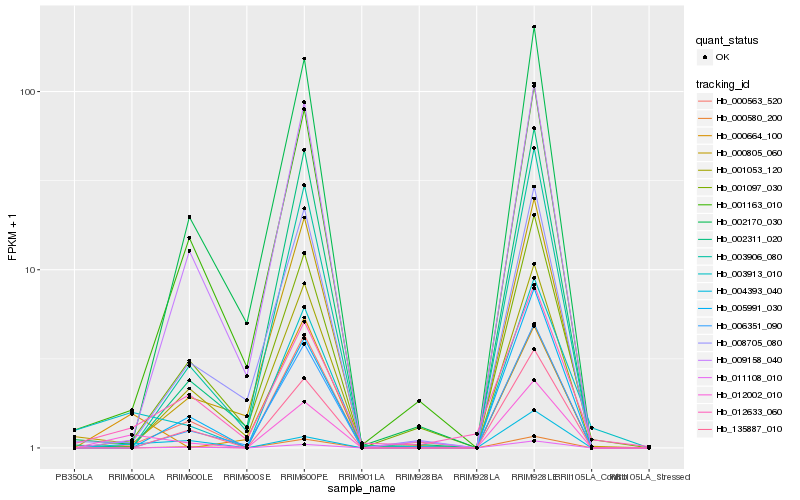
Hb_012002_010 |
0.0 |
- |
- |
hypothetical protein VITISV_016116 [Vitis vinifera] |
| 2 |
Hb_003913_010 |
0.1527333347 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000664_100 |
0.1960890336 |
- |
- |
PREDICTED: uncharacterized protein LOC105638386 [Jatropha curcas] |
| 4 |
Hb_000563_520 |
0.2003392053 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 5 |
Hb_003906_080 |
0.201362279 |
- |
- |
Peroxidase 66 precursor, putative [Ricinus communis] |
| 6 |
Hb_004393_040 |
0.2039671481 |
transcription factor |
TF Family: RAV |
PREDICTED: AP2/ERF and B3 domain-containing transcription factor At1g50680-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_005991_030 |
0.2063839172 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 8 |
Hb_012633_060 |
0.2066179916 |
- |
- |
laccase, putative [Ricinus communis] |
| 9 |
Hb_135887_010 |
0.2073264514 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 10 |
Hb_006351_090 |
0.2095455471 |
- |
- |
UDP-glycosyltransferase 85A1 [Gossypium arboreum] |
| 11 |
Hb_011108_010 |
0.2104068727 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103335582 [Prunus mume] |
| 12 |
Hb_000580_200 |
0.2104663483 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002311_020 |
0.2119803043 |
- |
- |
PREDICTED: putative beta-D-xylosidase [Jatropha curcas] |
| 14 |
Hb_002170_030 |
0.2131274594 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 15 |
Hb_000805_060 |
0.2139290823 |
- |
- |
laccase, putative [Ricinus communis] |
| 16 |
Hb_001097_030 |
0.214371783 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 17 |
Hb_001163_010 |
0.2153125546 |
- |
- |
PREDICTED: syncoilin [Jatropha curcas] |
| 18 |
Hb_008705_080 |
0.2161405703 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 19 |
Hb_001053_120 |
0.2169187188 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46-like [Populus euphratica] |
| 20 |
Hb_009158_040 |
0.2169347348 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |