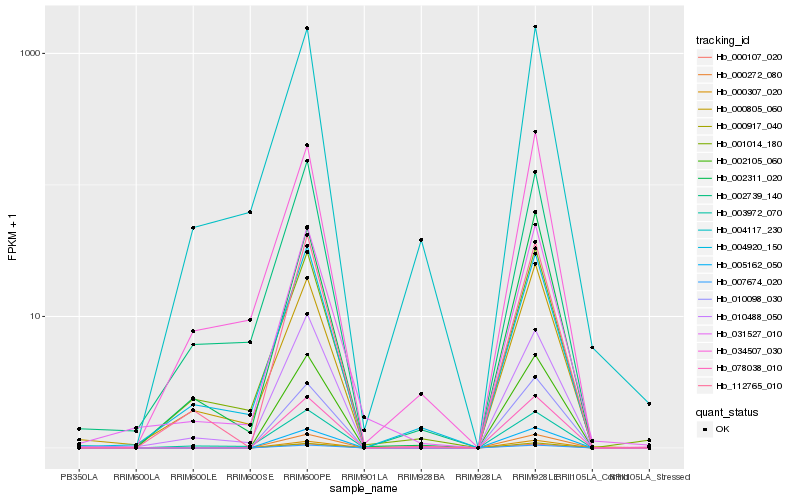
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031527_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002311_020 |
0.0987951683 |
- |
- |
PREDICTED: putative beta-D-xylosidase [Jatropha curcas] |
| 3 |
Hb_000805_060 |
0.1035245745 |
- |
- |
laccase, putative [Ricinus communis] |
| 4 |
Hb_001014_180 |
0.1037920719 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10 [Jatropha curcas] |
| 5 |
Hb_010488_050 |
0.1060121467 |
transcription factor |
TF Family: LIM |
hypothetical protein JCGZ_14428 [Jatropha curcas] |
| 6 |
Hb_112765_010 |
0.1079026874 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004920_150 |
0.1093309181 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003972_070 |
0.1099846417 |
- |
- |
PREDICTED: cytochrome P450 85A1-like [Citrus sinensis] |
| 9 |
Hb_034507_030 |
0.1129236459 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 10 |
Hb_002739_140 |
0.1140444226 |
- |
- |
PREDICTED: protein TPX2 [Jatropha curcas] |
| 11 |
Hb_078038_010 |
0.1146259261 |
- |
- |
hypothetical protein CISIN_1g038197mg [Citrus sinensis] |
| 12 |
Hb_005162_050 |
0.1148439627 |
- |
- |
hypothetical protein B456_009G352100 [Gossypium raimondii] |
| 13 |
Hb_002105_060 |
0.115040703 |
- |
- |
hypothetical protein POPTR_0022s00320g [Populus trichocarpa] |
| 14 |
Hb_000272_080 |
0.1151690649 |
- |
- |
temperature-induced lipocalin [Populus tremula x Populus tremuloides] |
| 15 |
Hb_007674_020 |
0.1154874453 |
- |
- |
PREDICTED: serpin-ZX-like [Jatropha curcas] |
| 16 |
Hb_000917_040 |
0.1157414441 |
- |
- |
PREDICTED: uncharacterized protein LOC105639996 [Jatropha curcas] |
| 17 |
Hb_000107_020 |
0.1168034944 |
- |
- |
PREDICTED: uncharacterized protein LOC105032542 [Elaeis guineensis] |
| 18 |
Hb_000307_020 |
0.1169197047 |
- |
- |
PREDICTED: uncharacterized protein LOC100255757 [Vitis vinifera] |
| 19 |
Hb_010098_030 |
0.1175066283 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004117_230 |
0.1177086787 |
- |
- |
lipid transfer precursor protein [Hevea brasiliensis] |