| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
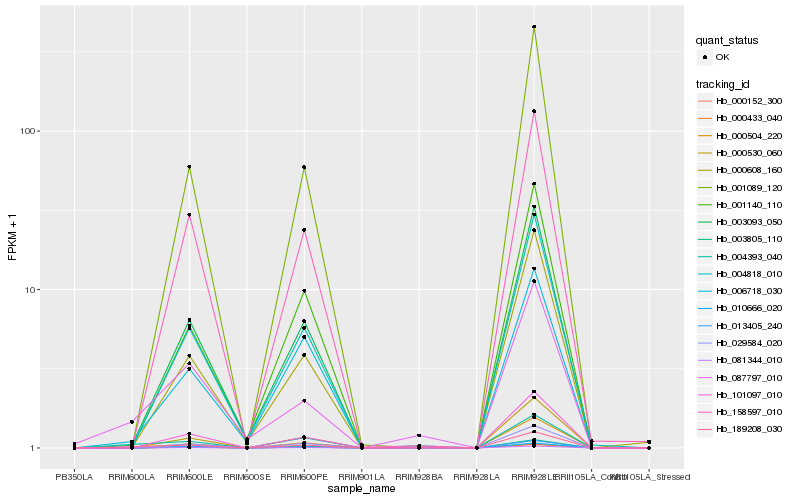
Hb_189208_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_004393_040 |
0.0711546193 |
transcription factor |
TF Family: RAV |
PREDICTED: AP2/ERF and B3 domain-containing transcription factor At1g50680-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_087797_010 |
0.1513600166 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_003093_050 |
0.1540011639 |
- |
- |
PREDICTED: uncharacterized protein LOC105641023 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000530_060 |
0.1578638216 |
- |
- |
- |
| 6 |
Hb_029584_020 |
0.1583702669 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001089_120 |
0.161550301 |
- |
- |
PREDICTED: pectinesterase [Jatropha curcas] |
| 8 |
Hb_003805_110 |
0.1620312388 |
- |
- |
PREDICTED: laccase-11-like [Jatropha curcas] |
| 9 |
Hb_081344_010 |
0.1626652454 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 10 |
Hb_000433_040 |
0.1634423913 |
- |
- |
- |
| 11 |
Hb_004818_010 |
0.1639405396 |
- |
- |
hypothetical protein JCGZ_13628 [Jatropha curcas] |
| 12 |
Hb_000504_220 |
0.1652632169 |
- |
- |
hypothetical protein JCGZ_10249 [Jatropha curcas] |
| 13 |
Hb_000152_300 |
0.1653748874 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 14 |
Hb_158597_010 |
0.1665266916 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial [Jatropha curcas] |
| 15 |
Hb_010666_020 |
0.1665819241 |
- |
- |
hypothetical protein JCGZ_04276 [Jatropha curcas] |
| 16 |
Hb_101097_010 |
0.1686432094 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 17 |
Hb_000608_160 |
0.1712305698 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 18 |
Hb_006718_030 |
0.1714796096 |
- |
- |
ATPase, F1 complex, gamma subunit protein [Theobroma cacao] |
| 19 |
Hb_001140_110 |
0.1715626701 |
- |
- |
hypothetical protein JCGZ_26123 [Jatropha curcas] |
| 20 |
Hb_013405_240 |
0.1716940286 |
- |
- |
Hydroxyproline-rich glycoprotein family protein [Theobroma cacao] |