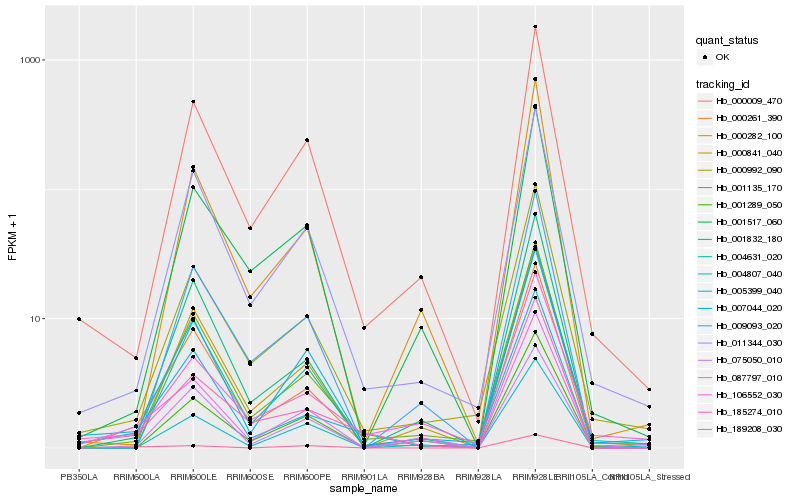
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_087797_010 |
0.0 |
- |
- |
kinase, putative [Ricinus communis] |
| 2 |
Hb_000261_390 |
0.1188736609 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 15 [Jatropha curcas] |
| 3 |
Hb_007044_020 |
0.1266275664 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_001832_180 |
0.1357248044 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein RCOM_1046780 [Ricinus communis] |
| 5 |
Hb_000009_470 |
0.13665831 |
- |
- |
Oxygen-evolving enhancer protein 1, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_001289_050 |
0.1384206239 |
- |
- |
hypothetical protein Poptr_cp075 [Populus trichocarpa] |
| 7 |
Hb_000282_100 |
0.1408297257 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 8 |
Hb_185274_010 |
0.1415645532 |
- |
- |
hypothetical protein ARALYDRAFT_330174 [Arabidopsis lyrata subsp. lyrata] |
| 9 |
Hb_001135_170 |
0.1431507843 |
transcription factor |
TF Family: Orphans |
Two-component response regulator ARR2, putative [Ricinus communis] |
| 10 |
Hb_000992_090 |
0.144050205 |
transcription factor |
TF Family: Orphans |
CIL, putative [Ricinus communis] |
| 11 |
Hb_106552_030 |
0.1444523624 |
- |
- |
hypothetical protein POPTR_0006s23180g [Populus trichocarpa] |
| 12 |
Hb_000841_040 |
0.1448608321 |
transcription factor |
TF Family: C2C2-CO-like |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_001517_060 |
0.1450265277 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |
| 14 |
Hb_004807_040 |
0.1464081634 |
- |
- |
PREDICTED: uncharacterized protein LOC104420559 [Eucalyptus grandis] |
| 15 |
Hb_075050_010 |
0.1469048002 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_009093_020 |
0.1472516702 |
- |
- |
PREDICTED: uncharacterized protein LOC105642100 [Jatropha curcas] |
| 17 |
Hb_005399_040 |
0.1476932348 |
- |
- |
RNA polymerase beta subunit [Hevea brasiliensis] |
| 18 |
Hb_011344_030 |
0.1486144555 |
- |
- |
PREDICTED: elongation factor G-2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_004631_020 |
0.1504946796 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 20 |
Hb_189208_030 |
0.1513600166 |
- |
- |
- |