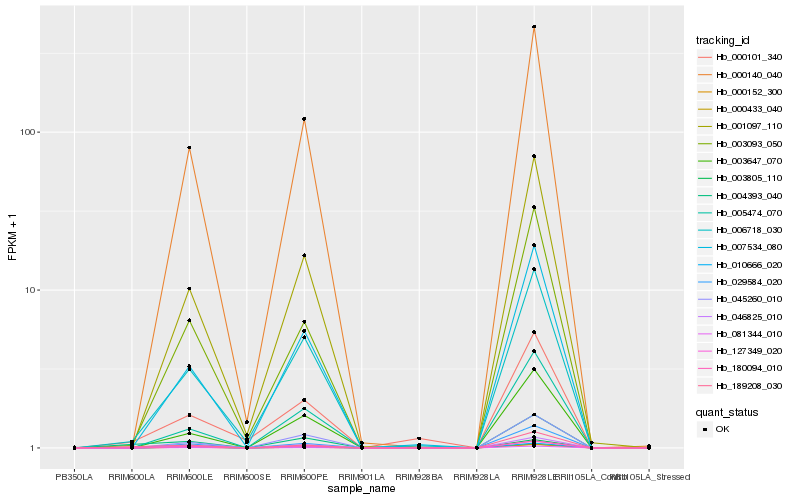
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004393_040 |
0.0 |
transcription factor |
TF Family: RAV |
PREDICTED: AP2/ERF and B3 domain-containing transcription factor At1g50680-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_189208_030 |
0.0711546193 |
- |
- |
- |
| 3 |
Hb_006718_030 |
0.1512926492 |
- |
- |
ATPase, F1 complex, gamma subunit protein [Theobroma cacao] |
| 4 |
Hb_000140_040 |
0.1710117763 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 5 |
Hb_180094_010 |
0.1733264706 |
- |
- |
PREDICTED: uncharacterized protein LOC105628311 [Jatropha curcas] |
| 6 |
Hb_003805_110 |
0.1751229356 |
- |
- |
PREDICTED: laccase-11-like [Jatropha curcas] |
| 7 |
Hb_000152_300 |
0.1751887102 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 8 |
Hb_127349_020 |
0.1752424441 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 9 |
Hb_007534_080 |
0.1759834836 |
- |
- |
d-3-phosphoglycerate dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_000101_340 |
0.1762004686 |
- |
- |
2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Ricinus communis] |
| 11 |
Hb_001097_110 |
0.1765390582 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 12 |
Hb_081344_010 |
0.1771053353 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 13 |
Hb_003647_070 |
0.1771472211 |
- |
- |
hypothetical protein POPTR_0006s09430g [Populus trichocarpa] |
| 14 |
Hb_000433_040 |
0.1774534593 |
- |
- |
- |
| 15 |
Hb_029584_020 |
0.1778260986 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_046825_010 |
0.177841988 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 17 |
Hb_005474_070 |
0.1778479836 |
- |
- |
hypothetical protein JCGZ_10579 [Jatropha curcas] |
| 18 |
Hb_010666_020 |
0.1782481878 |
- |
- |
hypothetical protein JCGZ_04276 [Jatropha curcas] |
| 19 |
Hb_003093_050 |
0.1790623208 |
- |
- |
PREDICTED: uncharacterized protein LOC105641023 isoform X2 [Jatropha curcas] |
| 20 |
Hb_045260_010 |
0.1794920352 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EMS1-like [Jatropha curcas] |