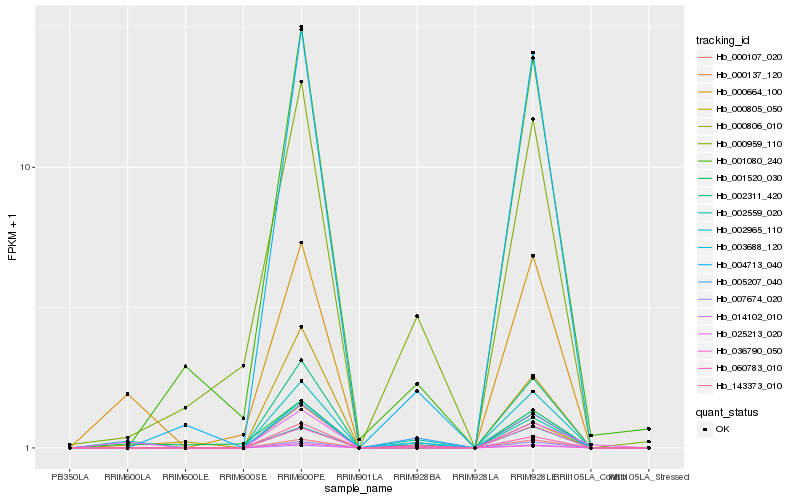
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002965_110 |
0.0 |
- |
- |
hypothetical protein POPTR_0010s15040g [Populus trichocarpa] |
| 2 |
Hb_000664_100 |
0.1545295701 |
- |
- |
PREDICTED: uncharacterized protein LOC105638386 [Jatropha curcas] |
| 3 |
Hb_004713_040 |
0.1618544704 |
- |
- |
hypothetical protein JCGZ_22567 [Jatropha curcas] |
| 4 |
Hb_025213_020 |
0.1635828314 |
- |
- |
PREDICTED: (R)-mandelonitrile lyase-like [Populus euphratica] |
| 5 |
Hb_000806_010 |
0.1749802654 |
- |
- |
PREDICTED: hevamine-A-like [Jatropha curcas] |
| 6 |
Hb_060783_010 |
0.1757698233 |
- |
- |
Chloride channel protein CLC-c [Populus trichocarpa] |
| 7 |
Hb_001520_030 |
0.1789958843 |
transcription factor |
TF Family: MYB |
PREDICTED: trichome differentiation protein GL1-like isoform X2 [Populus euphratica] |
| 8 |
Hb_036790_050 |
0.1817770683 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 9 |
Hb_002559_020 |
0.1818849485 |
transcription factor |
TF Family: TUB |
- |
| 10 |
Hb_005207_040 |
0.185543165 |
- |
- |
PREDICTED: zinc finger Ran-binding domain-containing protein 2-like [Cicer arietinum] |
| 11 |
Hb_014102_010 |
0.186072896 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 12 |
Hb_000805_050 |
0.188995056 |
- |
- |
zinc transporter, putative [Ricinus communis] |
| 13 |
Hb_000137_120 |
0.1911532977 |
- |
- |
hypothetical protein JCGZ_15021 [Jatropha curcas] |
| 14 |
Hb_143373_010 |
0.1915494281 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 [Gossypium raimondii] |
| 15 |
Hb_002311_420 |
0.1917309318 |
- |
- |
hypothetical protein JCGZ_18919 [Jatropha curcas] |
| 16 |
Hb_003688_120 |
0.1919537991 |
- |
- |
PREDICTED: uncharacterized protein LOC105350793 [Fragaria vesca subsp. vesca] |
| 17 |
Hb_000959_110 |
0.1924238613 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 13 [Vitis vinifera] |
| 18 |
Hb_001080_240 |
0.1931147813 |
- |
- |
hypothetical protein PHAVU_001G152800g [Phaseolus vulgaris] |
| 19 |
Hb_000107_020 |
0.1932266339 |
- |
- |
PREDICTED: uncharacterized protein LOC105032542 [Elaeis guineensis] |
| 20 |
Hb_007674_020 |
0.1947079558 |
- |
- |
PREDICTED: serpin-ZX-like [Jatropha curcas] |