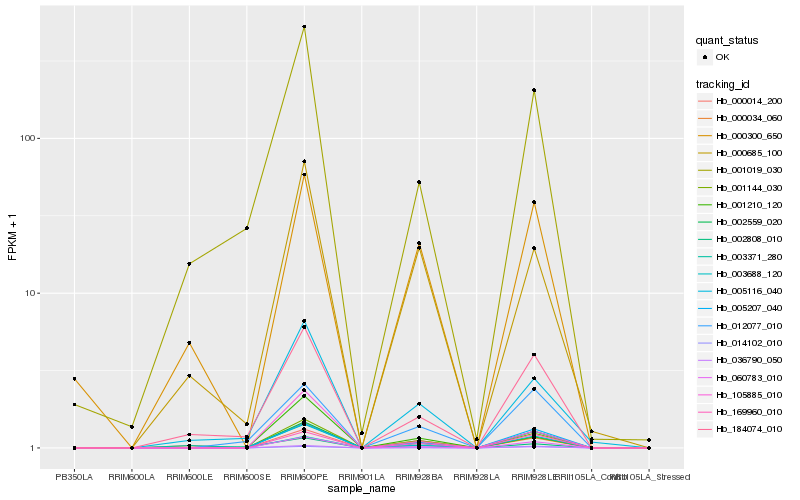
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001144_030 |
0.0 |
- |
- |
hypothetical protein CICLE_v10023280mg, partial [Citrus clementina] |
| 2 |
Hb_000014_200 |
0.0777378534 |
- |
- |
PREDICTED: uncharacterized protein LOC105775355 [Gossypium raimondii] |
| 3 |
Hb_002559_020 |
0.0796266047 |
transcription factor |
TF Family: TUB |
- |
| 4 |
Hb_060783_010 |
0.0963331815 |
- |
- |
Chloride channel protein CLC-c [Populus trichocarpa] |
| 5 |
Hb_005207_040 |
0.1013196467 |
- |
- |
PREDICTED: zinc finger Ran-binding domain-containing protein 2-like [Cicer arietinum] |
| 6 |
Hb_036790_050 |
0.1060878932 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 7 |
Hb_169960_010 |
0.1198219675 |
- |
- |
- |
| 8 |
Hb_000685_100 |
0.1535786596 |
- |
- |
hypothetical protein POPTR_0001s10630g [Populus trichocarpa] |
| 9 |
Hb_005116_040 |
0.1589551581 |
- |
- |
PREDICTED: uncharacterized protein LOC105131445 [Populus euphratica] |
| 10 |
Hb_000034_060 |
0.1606492481 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Citrus sinensis] |
| 11 |
Hb_003688_120 |
0.1617523089 |
- |
- |
PREDICTED: uncharacterized protein LOC105350793 [Fragaria vesca subsp. vesca] |
| 12 |
Hb_000300_650 |
0.1757807025 |
- |
- |
PREDICTED: arabinogalactan peptide 13-like [Jatropha curcas] |
| 13 |
Hb_012077_010 |
0.1768854996 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 14 |
Hb_001210_120 |
0.1774224707 |
- |
- |
hypothetical protein JCGZ_06226 [Jatropha curcas] |
| 15 |
Hb_002808_010 |
0.1793153806 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 16 |
Hb_014102_010 |
0.1794139619 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 17 |
Hb_184074_010 |
0.17991527 |
- |
- |
PREDICTED: uncharacterized protein LOC105639054 [Jatropha curcas] |
| 18 |
Hb_001019_030 |
0.186220401 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 19 |
Hb_105885_010 |
0.1903491512 |
- |
- |
PREDICTED: uncharacterized protein LOC103441690 [Malus domestica] |
| 20 |
Hb_003371_280 |
0.1914501418 |
- |
- |
unnamed protein product [Vitis vinifera] |