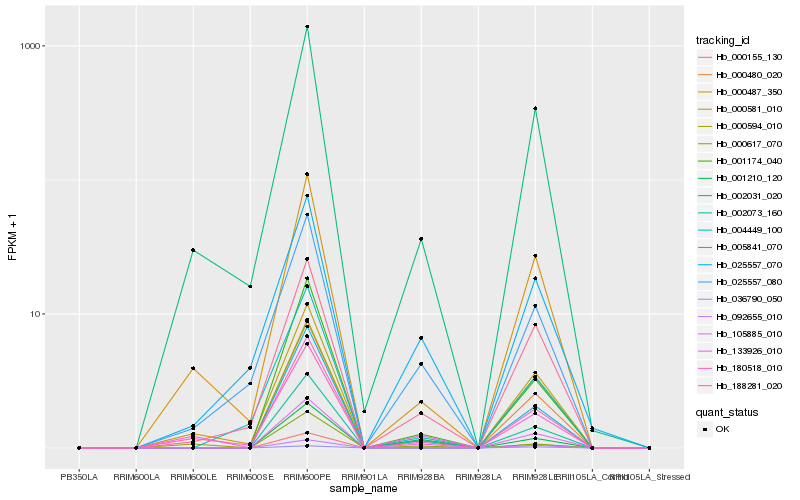
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_105885_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103441690 [Malus domestica] |
| 2 |
Hb_002073_160 |
0.0322383385 |
- |
- |
hypothetical protein JCGZ_21740 [Jatropha curcas] |
| 3 |
Hb_001174_040 |
0.1021269906 |
- |
- |
- |
| 4 |
Hb_188281_020 |
0.1136744958 |
- |
- |
PREDICTED: uncharacterized protein LOC105629522 [Jatropha curcas] |
| 5 |
Hb_001210_120 |
0.1143454343 |
- |
- |
hypothetical protein JCGZ_06226 [Jatropha curcas] |
| 6 |
Hb_002031_020 |
0.126193447 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 7 |
Hb_025557_080 |
0.1332643635 |
transcription factor |
TF Family: zf-HD |
- |
| 8 |
Hb_133926_010 |
0.1400894855 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 9 |
Hb_025557_070 |
0.1421720028 |
transcription factor |
TF Family: zf-HD |
hypothetical protein JCGZ_11626 [Jatropha curcas] |
| 10 |
Hb_000581_010 |
0.1425677519 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6-like [Jatropha curcas] |
| 11 |
Hb_000594_010 |
0.1439861297 |
- |
- |
PREDICTED: lysine histidine transporter-like 6 isoform X3 [Populus euphratica] |
| 12 |
Hb_000155_130 |
0.1443086834 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000480_020 |
0.144521299 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000617_070 |
0.1452519707 |
- |
- |
hypothetical protein POPTR_0005s02420g [Populus trichocarpa] |
| 15 |
Hb_000487_350 |
0.1454992251 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_180518_010 |
0.1484440399 |
- |
- |
PREDICTED: receptor-like protein 2 [Jatropha curcas] |
| 17 |
Hb_004449_100 |
0.1508883487 |
- |
- |
- |
| 18 |
Hb_036790_050 |
0.1520431259 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 19 |
Hb_005841_070 |
0.1528792201 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_092655_010 |
0.1554334727 |
- |
- |
PREDICTED: uncharacterized protein LOC105650151 [Jatropha curcas] |