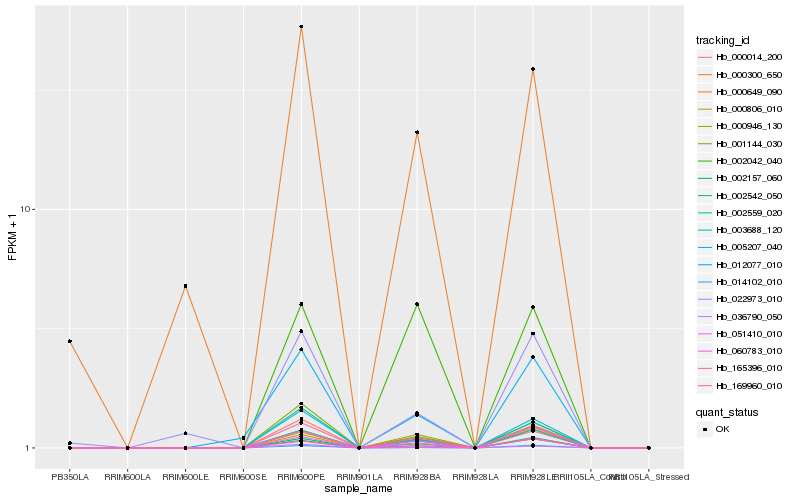
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_169960_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000014_200 |
0.0755916097 |
- |
- |
PREDICTED: uncharacterized protein LOC105775355 [Gossypium raimondii] |
| 3 |
Hb_005207_040 |
0.0894750576 |
- |
- |
PREDICTED: zinc finger Ran-binding domain-containing protein 2-like [Cicer arietinum] |
| 4 |
Hb_003688_120 |
0.0981508876 |
- |
- |
PREDICTED: uncharacterized protein LOC105350793 [Fragaria vesca subsp. vesca] |
| 5 |
Hb_014102_010 |
0.1197163171 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 6 |
Hb_001144_030 |
0.1198219675 |
- |
- |
hypothetical protein CICLE_v10023280mg, partial [Citrus clementina] |
| 7 |
Hb_002559_020 |
0.1253366238 |
transcription factor |
TF Family: TUB |
- |
| 8 |
Hb_000649_090 |
0.1325530032 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_012077_010 |
0.1416549088 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 10 |
Hb_002157_060 |
0.1493755569 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 9 [Jatropha curcas] |
| 11 |
Hb_000946_130 |
0.1550631957 |
- |
- |
PREDICTED: uncharacterized protein LOC104602430 [Nelumbo nucifera] |
| 12 |
Hb_000300_650 |
0.1583442882 |
- |
- |
PREDICTED: arabinogalactan peptide 13-like [Jatropha curcas] |
| 13 |
Hb_060783_010 |
0.1589638107 |
- |
- |
Chloride channel protein CLC-c [Populus trichocarpa] |
| 14 |
Hb_002042_040 |
0.1666451533 |
- |
- |
hypothetical protein POPTR_0013s02440g [Populus trichocarpa] |
| 15 |
Hb_165396_010 |
0.1733056403 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 16 |
Hb_002542_050 |
0.177828056 |
- |
- |
- |
| 17 |
Hb_000806_010 |
0.1824584726 |
- |
- |
PREDICTED: hevamine-A-like [Jatropha curcas] |
| 18 |
Hb_022973_010 |
0.1826923655 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Jatropha curcas] |
| 19 |
Hb_036790_050 |
0.1829908728 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 20 |
Hb_051410_010 |
0.189302865 |
- |
- |
PREDICTED: uncharacterized protein LOC103401813 [Malus domestica] |