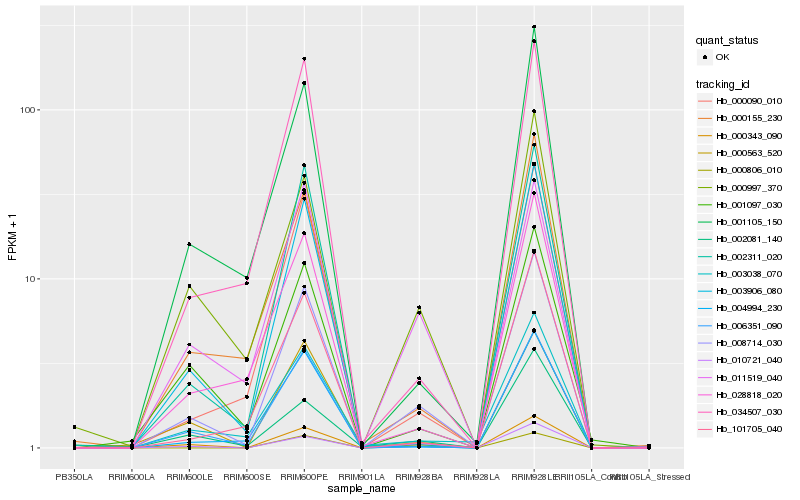
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008714_030 |
0.0 |
- |
- |
Multidrug resistance protein ABC transporter family protein, putative [Theobroma cacao] |
| 2 |
Hb_000343_090 |
0.0730009144 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000090_010 |
0.1202957998 |
- |
- |
hypothetical protein JCGZ_22567 [Jatropha curcas] |
| 4 |
Hb_006351_090 |
0.121850256 |
- |
- |
UDP-glycosyltransferase 85A1 [Gossypium arboreum] |
| 5 |
Hb_000997_370 |
0.1252101283 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Jatropha curcas] |
| 6 |
Hb_003906_080 |
0.1291734624 |
- |
- |
Peroxidase 66 precursor, putative [Ricinus communis] |
| 7 |
Hb_010721_040 |
0.1303461894 |
transcription factor |
TF Family: M-type |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_000806_010 |
0.1316248874 |
- |
- |
PREDICTED: hevamine-A-like [Jatropha curcas] |
| 9 |
Hb_001097_030 |
0.1335085163 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 10 |
Hb_000155_230 |
0.1345312061 |
- |
- |
rac gtpase, putative [Ricinus communis] |
| 11 |
Hb_000563_520 |
0.1348806604 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 12 |
Hb_004994_230 |
0.1351111563 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3, putative [Ricinus communis] |
| 13 |
Hb_003038_070 |
0.1367356274 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 14 |
Hb_028818_020 |
0.1386680329 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9-like [Jatropha curcas] |
| 15 |
Hb_002311_020 |
0.1399336242 |
- |
- |
PREDICTED: putative beta-D-xylosidase [Jatropha curcas] |
| 16 |
Hb_101705_040 |
0.1405609084 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_011519_040 |
0.1405755196 |
- |
- |
hypothetical protein POPTR_0015s04770g [Populus trichocarpa] |
| 18 |
Hb_002081_140 |
0.1411095169 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001105_150 |
0.142274179 |
- |
- |
GERMIN-LIKE protein 10 [Populus trichocarpa] |
| 20 |
Hb_034507_030 |
0.1424532167 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |