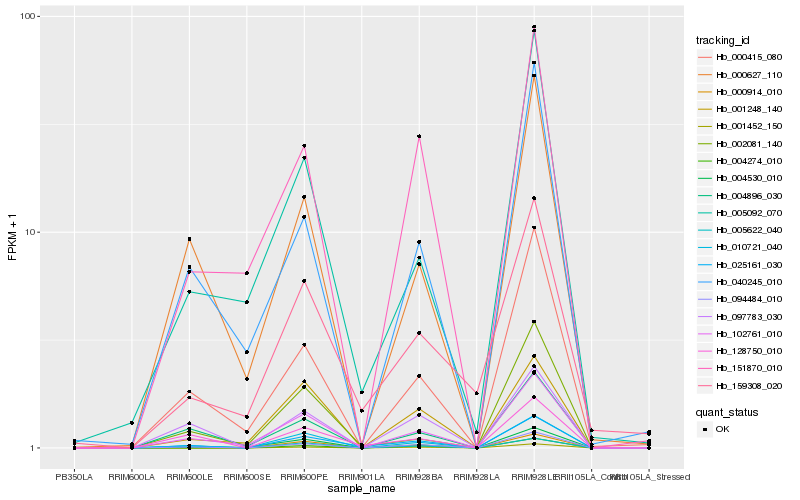
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025161_030 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 2 |
Hb_094484_010 |
0.1199454009 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_010721_040 |
0.1274627467 |
transcription factor |
TF Family: M-type |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_128750_010 |
0.1425558529 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 5 |
Hb_102761_010 |
0.1457995855 |
- |
- |
hypothetical protein JCGZ_21614 [Jatropha curcas] |
| 6 |
Hb_004530_010 |
0.1692248238 |
- |
- |
PREDICTED: uncharacterized protein LOC104898079 [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_000415_080 |
0.1721372712 |
- |
- |
- |
| 8 |
Hb_097783_030 |
0.1787403326 |
- |
- |
PREDICTED: probable peroxidase 61 [Jatropha curcas] |
| 9 |
Hb_004896_030 |
0.1809922921 |
- |
- |
hypothetical protein VITISV_007475 [Vitis vinifera] |
| 10 |
Hb_005092_070 |
0.1824249164 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 11 |
Hb_005622_040 |
0.1844566334 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 12 |
Hb_004274_010 |
0.1852858014 |
- |
- |
PREDICTED: uncharacterized protein LOC104451199 [Eucalyptus grandis] |
| 13 |
Hb_000627_110 |
0.185605101 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 14 |
Hb_001248_140 |
0.1879814815 |
- |
- |
Thylakoid lumenal 19 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_000914_010 |
0.1880552103 |
- |
- |
hypothetical protein POPTR_0025s00760g [Populus trichocarpa] |
| 16 |
Hb_040245_010 |
0.1893165456 |
- |
- |
hypothetical protein CISIN_1g039575mg, partial [Citrus sinensis] |
| 17 |
Hb_159308_020 |
0.189365316 |
- |
- |
hypothetical protein B456_002G027900 [Gossypium raimondii] |
| 18 |
Hb_002081_140 |
0.1897929706 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001452_150 |
0.190395134 |
- |
- |
PREDICTED: uncharacterized protein LOC104879858 [Vitis vinifera] |
| 20 |
Hb_151870_010 |
0.1910457338 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |