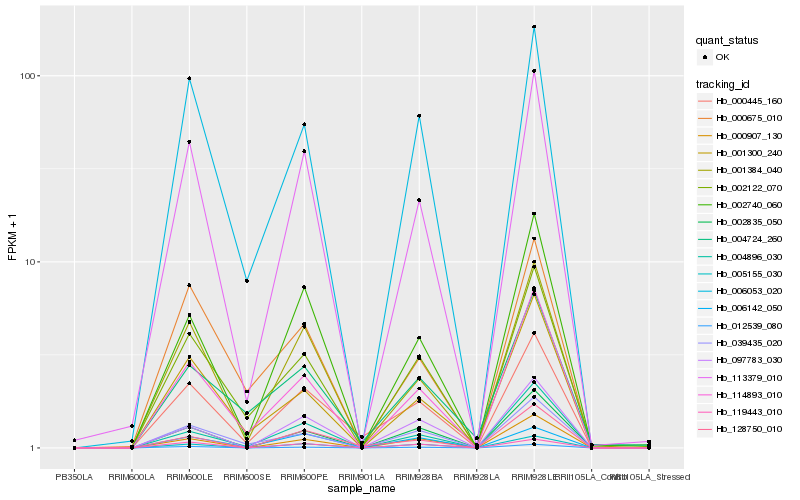
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000445_160 |
0.0 |
- |
- |
PREDICTED: probable calcium-binding protein CML44 [Jatropha curcas] |
| 2 |
Hb_113379_010 |
0.1191326268 |
- |
- |
PREDICTED: beta-xylosidase/alpha-L-arabinofuranosidase 1 [Jatropha curcas] |
| 3 |
Hb_128750_010 |
0.1249663702 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 4 |
Hb_001384_040 |
0.1249801608 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 5 |
Hb_114893_010 |
0.1309762019 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 6 |
Hb_006142_050 |
0.1311953267 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 7 |
Hb_006053_020 |
0.1322578739 |
- |
- |
CAP [Theobroma cacao] |
| 8 |
Hb_005155_030 |
0.1359799498 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 9 |
Hb_002122_070 |
0.1360674643 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_002740_060 |
0.1383607451 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 11 |
Hb_039435_020 |
0.1402029692 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 12 |
Hb_002835_050 |
0.1485998794 |
- |
- |
PREDICTED: high-affinity nitrate transporter 3.1-like [Jatropha curcas] |
| 13 |
Hb_012539_080 |
0.1494685301 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 14 |
Hb_004724_260 |
0.1562217341 |
- |
- |
PREDICTED: 40S ribosomal protein S14-2-like [Gossypium raimondii] |
| 15 |
Hb_000675_010 |
0.1565165159 |
- |
- |
PREDICTED: cytokinin dehydrogenase 7 [Jatropha curcas] |
| 16 |
Hb_000907_130 |
0.1566446211 |
- |
- |
- |
| 17 |
Hb_001300_240 |
0.1590053509 |
- |
- |
shikimate dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_119443_010 |
0.1595993916 |
- |
- |
hypothetical protein JCGZ_17348 [Jatropha curcas] |
| 19 |
Hb_004896_030 |
0.1601020428 |
- |
- |
hypothetical protein VITISV_007475 [Vitis vinifera] |
| 20 |
Hb_097783_030 |
0.160324958 |
- |
- |
PREDICTED: probable peroxidase 61 [Jatropha curcas] |