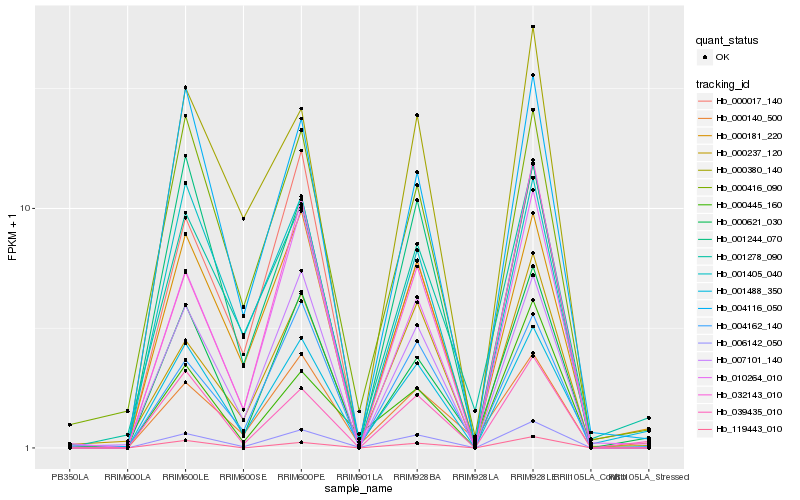
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006142_050 |
0.0 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 2 |
Hb_000237_120 |
0.105785807 |
- |
- |
PREDICTED: beta-galactosidase 3-like [Jatropha curcas] |
| 3 |
Hb_032143_010 |
0.1097963959 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 4 |
Hb_007101_140 |
0.1110286522 |
- |
- |
hypothetical protein JCGZ_21777 [Jatropha curcas] |
| 5 |
Hb_010264_010 |
0.120620118 |
- |
- |
PREDICTED: protein ZINC INDUCED FACILITATOR-LIKE 1-like [Jatropha curcas] |
| 6 |
Hb_000621_030 |
0.1268984871 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004162_140 |
0.1301595084 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 8 |
Hb_004116_050 |
0.1304701469 |
- |
- |
PREDICTED: cytochrome P450 90A1 [Jatropha curcas] |
| 9 |
Hb_000445_160 |
0.1311953267 |
- |
- |
PREDICTED: probable calcium-binding protein CML44 [Jatropha curcas] |
| 10 |
Hb_000380_140 |
0.1319454342 |
- |
- |
PREDICTED: uncharacterized protein LOC105628069 [Jatropha curcas] |
| 11 |
Hb_119443_010 |
0.1320759402 |
- |
- |
hypothetical protein JCGZ_17348 [Jatropha curcas] |
| 12 |
Hb_000181_220 |
0.1382501798 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_001488_350 |
0.139809645 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001405_040 |
0.139854773 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Jatropha curcas] |
| 15 |
Hb_000416_090 |
0.1413148275 |
- |
- |
Aquaporin NIP1.1, putative [Ricinus communis] |
| 16 |
Hb_039435_010 |
0.1416526419 |
- |
- |
hypothetical protein SORBIDRAFT_10g025530 [Sorghum bicolor] |
| 17 |
Hb_000017_140 |
0.1425939892 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 18 |
Hb_000140_500 |
0.1431993341 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_001244_070 |
0.1438389176 |
- |
- |
hypothetical protein POPTR_0004s13470g [Populus trichocarpa] |
| 20 |
Hb_001278_090 |
0.1439499666 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |