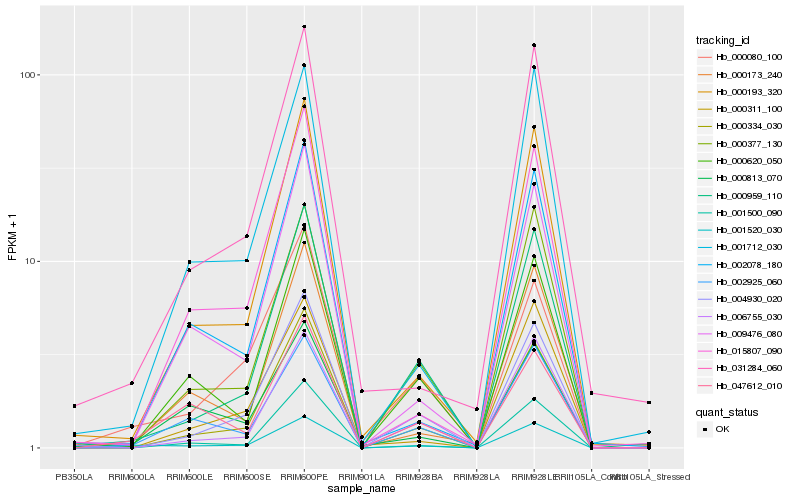
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001520_030 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: trichome differentiation protein GL1-like isoform X2 [Populus euphratica] |
| 2 |
Hb_002925_060 |
0.1360779288 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 3 |
Hb_000959_110 |
0.1420682863 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 13 [Vitis vinifera] |
| 4 |
Hb_000193_320 |
0.143116591 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000377_130 |
0.1444651213 |
- |
- |
hypothetical protein POPTR_0016s00790g [Populus trichocarpa] |
| 6 |
Hb_000334_030 |
0.1448125389 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001712_030 |
0.15008495 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 19 [Jatropha curcas] |
| 8 |
Hb_001500_090 |
0.1543223054 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_009476_080 |
0.1555341703 |
- |
- |
BnaA03g18920D [Brassica napus] |
| 10 |
Hb_031284_060 |
0.1567693931 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 7 [UDP-forming] [Jatropha curcas] |
| 11 |
Hb_004930_020 |
0.1576682336 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 12 |
Hb_000080_100 |
0.1577102903 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 13 |
Hb_047612_010 |
0.1593467209 |
- |
- |
glutamine amidotransferase class-I domain-containing family protein [Populus trichocarpa] |
| 14 |
Hb_002078_180 |
0.1600817586 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 8 [Jatropha curcas] |
| 15 |
Hb_006755_030 |
0.1602919647 |
- |
- |
BURP domain-containing protein, putative [Theobroma cacao] |
| 16 |
Hb_000173_240 |
0.1603001801 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000620_050 |
0.1612754194 |
- |
- |
PREDICTED: uncharacterized protein LOC105649924 [Jatropha curcas] |
| 18 |
Hb_000311_100 |
0.1615173038 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000813_070 |
0.1619863402 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_015807_090 |
0.1628330402 |
- |
- |
PREDICTED: probable galacturonosyltransferase 12 [Jatropha curcas] |