| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
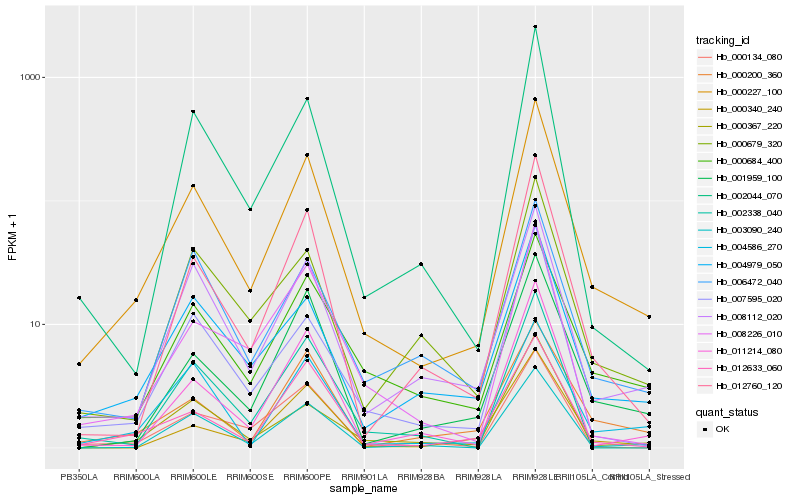
Hb_000227_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631933 [Jatropha curcas] |
| 2 |
Hb_012760_120 |
0.1150386294 |
- |
- |
PREDICTED: peroxidase 64-like [Jatropha curcas] |
| 3 |
Hb_008112_020 |
0.1157383315 |
- |
- |
PREDICTED: magnesium transporter MRS2-11, chloroplastic [Jatropha curcas] |
| 4 |
Hb_004979_050 |
0.1176776278 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 5 |
Hb_003090_240 |
0.1246653598 |
- |
- |
hypothetical protein JCGZ_25095 [Jatropha curcas] |
| 6 |
Hb_001959_100 |
0.1274597416 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 7 |
Hb_000684_400 |
0.1287275878 |
- |
- |
PREDICTED: uncharacterized protein LOC105642159 [Jatropha curcas] |
| 8 |
Hb_002044_070 |
0.1324478165 |
- |
- |
Chlorophyll a-b binding protein 3, chloroplastic [Theobroma cacao] |
| 9 |
Hb_004586_270 |
0.13341914 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_011214_080 |
0.1343042747 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 11 |
Hb_012633_060 |
0.1369699169 |
- |
- |
laccase, putative [Ricinus communis] |
| 12 |
Hb_000367_220 |
0.1384241114 |
- |
- |
hypothetical protein RCOM_1470580 [Ricinus communis] |
| 13 |
Hb_000679_320 |
0.1398346809 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105136146 [Populus euphratica] |
| 14 |
Hb_000200_360 |
0.1412272554 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 15 |
Hb_006472_040 |
0.1459506818 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000134_080 |
0.1461898517 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 17 |
Hb_008226_010 |
0.147650801 |
- |
- |
PREDICTED: uncharacterized protein LOC105649179 [Jatropha curcas] |
| 18 |
Hb_007595_020 |
0.1477435773 |
- |
- |
maturase K [Hevea brasiliensis] |
| 19 |
Hb_002338_040 |
0.1481051828 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000340_240 |
0.1502841203 |
- |
- |
conserved hypothetical protein [Ricinus communis] |