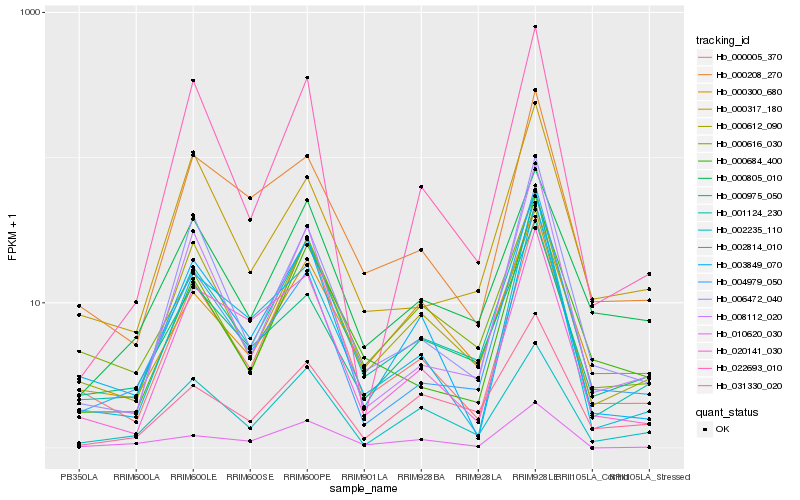
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001124_230 |
0.0 |
- |
- |
PREDICTED: divinyl chlorophyllide a 8-vinyl-reductase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000616_030 |
0.1044313321 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 3 |
Hb_000005_370 |
0.1127760695 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105634024 [Jatropha curcas] |
| 4 |
Hb_000612_090 |
0.1211291647 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 5 |
Hb_008112_020 |
0.1215582942 |
- |
- |
PREDICTED: magnesium transporter MRS2-11, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000300_680 |
0.1267844052 |
- |
- |
PREDICTED: uncharacterized protein LOC105632625 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000684_400 |
0.1268252713 |
- |
- |
PREDICTED: uncharacterized protein LOC105642159 [Jatropha curcas] |
| 8 |
Hb_006472_040 |
0.1290751837 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_010620_030 |
0.130853851 |
- |
- |
beta-amyrin synthase [Euphorbia tirucalli] |
| 10 |
Hb_002235_110 |
0.1312824043 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 11 |
Hb_000208_270 |
0.1317673489 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 12 |
Hb_000975_050 |
0.1323218675 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003849_070 |
0.1348271329 |
- |
- |
PREDICTED: uncharacterized protein LOC105644690 [Jatropha curcas] |
| 14 |
Hb_000805_010 |
0.1359066165 |
- |
- |
PREDICTED: uncharacterized protein LOC105629931 [Jatropha curcas] |
| 15 |
Hb_022693_010 |
0.1383461221 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 16 |
Hb_002814_010 |
0.1391102584 |
- |
- |
PREDICTED: adenylate kinase 5, chloroplastic [Jatropha curcas] |
| 17 |
Hb_031330_020 |
0.1402466501 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 18 |
Hb_000317_180 |
0.1403111256 |
- |
- |
PREDICTED: protein THYLAKOID FORMATION1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_020141_030 |
0.1404638672 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 20 |
Hb_004979_050 |
0.1405442231 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |