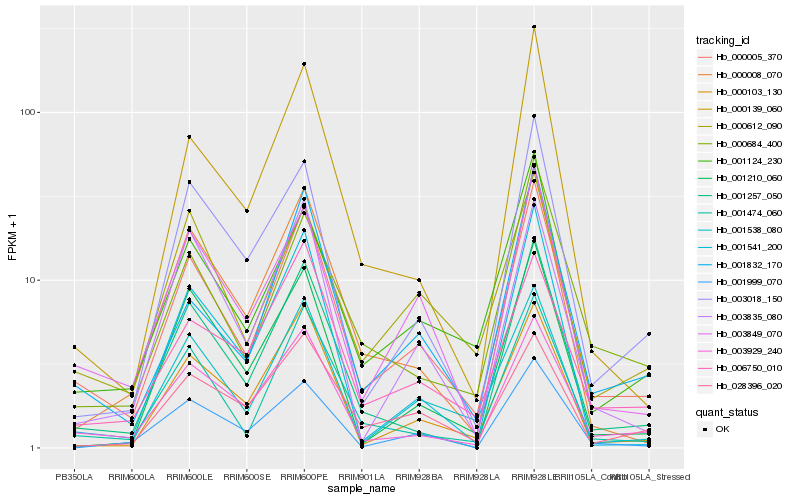
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000005_370 |
0.0 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105634024 [Jatropha curcas] |
| 2 |
Hb_003929_240 |
0.1021004735 |
- |
- |
PREDICTED: uncharacterized protein LOC105643616 isoform X2 [Jatropha curcas] |
| 3 |
Hb_003849_070 |
0.1079914774 |
- |
- |
PREDICTED: uncharacterized protein LOC105644690 [Jatropha curcas] |
| 4 |
Hb_001474_060 |
0.1113948317 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 16 [Jatropha curcas] |
| 5 |
Hb_001124_230 |
0.1127760695 |
- |
- |
PREDICTED: divinyl chlorophyllide a 8-vinyl-reductase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001832_170 |
0.1149478351 |
- |
- |
PREDICTED: beta-galactosidase-like [Jatropha curcas] |
| 7 |
Hb_001257_050 |
0.1170879055 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: protein YLS9 [Jatropha curcas] |
| 8 |
Hb_000103_130 |
0.1285471357 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 9 |
Hb_000684_400 |
0.1311623597 |
- |
- |
PREDICTED: uncharacterized protein LOC105642159 [Jatropha curcas] |
| 10 |
Hb_000008_070 |
0.1316699552 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_001538_080 |
0.131801662 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000139_060 |
0.132562043 |
- |
- |
Photosystem I reaction center subunit II, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_003018_150 |
0.1327448538 |
- |
- |
PREDICTED: uncharacterized protein LOC105628869 [Jatropha curcas] |
| 14 |
Hb_000612_090 |
0.1336330934 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 15 |
Hb_006750_010 |
0.1340448112 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 16 |
Hb_001999_070 |
0.1414431406 |
- |
- |
PREDICTED: ammonium transporter 1 member 3 [Jatropha curcas] |
| 17 |
Hb_001210_060 |
0.1431534744 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 18 |
Hb_028396_020 |
0.1438329471 |
- |
- |
PREDICTED: ras-related protein Rab7 [Jatropha curcas] |
| 19 |
Hb_001541_200 |
0.1442693527 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 20 |
Hb_003835_080 |
0.1452274575 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |