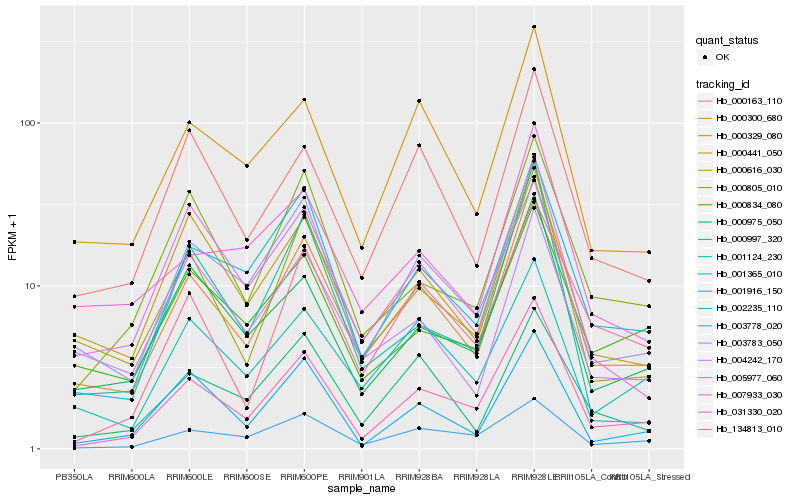
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_680 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632625 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000616_030 |
0.0872239224 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 3 |
Hb_031330_020 |
0.0942404944 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 4 |
Hb_000329_080 |
0.1062793428 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001916_150 |
0.1149545256 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-3 [Vitis vinifera] |
| 6 |
Hb_000163_110 |
0.1176492138 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 7 |
Hb_000975_050 |
0.1178323684 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001365_010 |
0.1230687443 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 9 |
Hb_000441_050 |
0.1238087569 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 10 |
Hb_001124_230 |
0.1267844052 |
- |
- |
PREDICTED: divinyl chlorophyllide a 8-vinyl-reductase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_007933_030 |
0.1277406166 |
- |
- |
PREDICTED: uncharacterized protein LOC105639932 [Jatropha curcas] |
| 12 |
Hb_005977_060 |
0.1300465895 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000997_320 |
0.1339651379 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme-like [Malus domestica] |
| 14 |
Hb_000834_080 |
0.1384998709 |
- |
- |
PREDICTED: uncharacterized protein LOC105637718 [Jatropha curcas] |
| 15 |
Hb_003783_050 |
0.1399101338 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000805_010 |
0.1403792519 |
- |
- |
PREDICTED: uncharacterized protein LOC105629931 [Jatropha curcas] |
| 17 |
Hb_004242_170 |
0.141324509 |
- |
- |
calcium/calmodulin-regulated receptor-like kinase [Manihot esculenta] |
| 18 |
Hb_002235_110 |
0.1458195976 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 19 |
Hb_134813_010 |
0.1458347527 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 20 |
Hb_003778_020 |
0.1477162624 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |