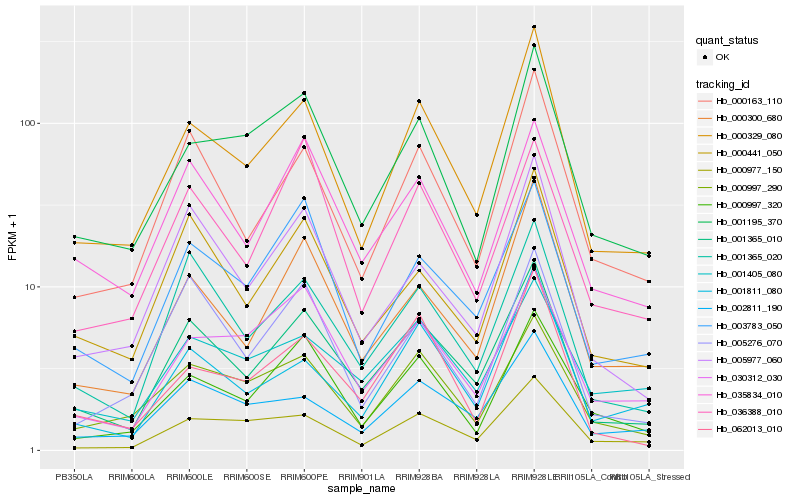
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001365_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 2 |
Hb_001365_020 |
0.0875499912 |
- |
- |
hypothetical protein PRUPE_ppa000927mg [Prunus persica] |
| 3 |
Hb_000329_080 |
0.0939947434 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000163_110 |
0.1059668492 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 5 |
Hb_000441_050 |
0.1119036597 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 6 |
Hb_003783_050 |
0.1170256297 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_035834_010 |
0.1197127095 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_005977_060 |
0.1201364454 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000300_680 |
0.1230687443 |
- |
- |
PREDICTED: uncharacterized protein LOC105632625 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002811_190 |
0.1255697787 |
- |
- |
tRNA pseudouridine synthase d, putative [Ricinus communis] |
| 11 |
Hb_000997_290 |
0.1330810109 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 12 |
Hb_000997_320 |
0.1379182015 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme-like [Malus domestica] |
| 13 |
Hb_001195_370 |
0.1393764384 |
- |
- |
- |
| 14 |
Hb_005276_070 |
0.1409481414 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 15 |
Hb_001405_080 |
0.1419912885 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 16 |
Hb_001811_080 |
0.1433769844 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 17 |
Hb_030312_030 |
0.1436692997 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 18 |
Hb_062013_010 |
0.1445940696 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 19 |
Hb_000977_150 |
0.144833172 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 20 |
Hb_036388_010 |
0.1461865416 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |