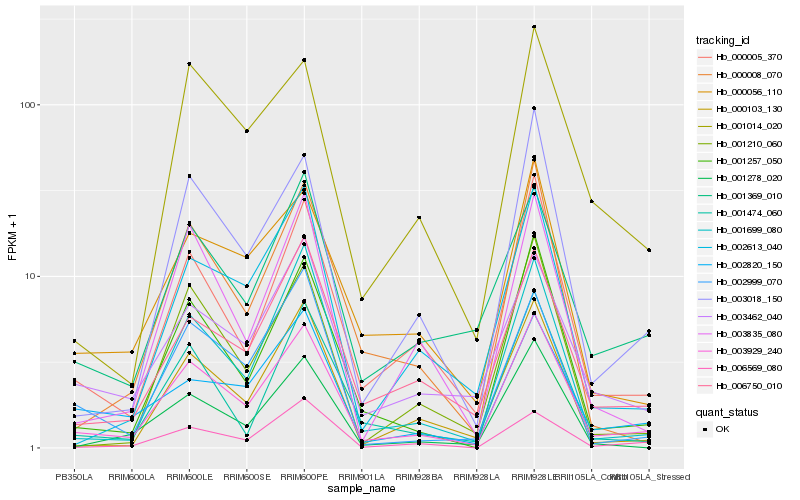
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003929_240 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643616 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001257_050 |
0.1010239342 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: protein YLS9 [Jatropha curcas] |
| 3 |
Hb_000005_370 |
0.1021004735 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105634024 [Jatropha curcas] |
| 4 |
Hb_003018_150 |
0.1214563224 |
- |
- |
PREDICTED: uncharacterized protein LOC105628869 [Jatropha curcas] |
| 5 |
Hb_006750_010 |
0.1260053071 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 6 |
Hb_001699_080 |
0.1280032412 |
- |
- |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_000103_130 |
0.1305751285 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 8 |
Hb_000056_110 |
0.1356553179 |
- |
- |
PREDICTED: uncharacterized protein LOC105630405 [Jatropha curcas] |
| 9 |
Hb_000008_070 |
0.1364878087 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_003462_040 |
0.1370935997 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_001210_060 |
0.1394624991 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 12 |
Hb_002613_040 |
0.1404040188 |
- |
- |
phosphate translocator-related family protein [Populus trichocarpa] |
| 13 |
Hb_002820_150 |
0.1412335398 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_003835_080 |
0.1416119536 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001278_020 |
0.1422511678 |
- |
- |
sucrose synthase 6 [Hevea brasiliensis] |
| 16 |
Hb_001474_060 |
0.1432421238 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 16 [Jatropha curcas] |
| 17 |
Hb_001369_010 |
0.1441453463 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 18 |
Hb_006569_080 |
0.1446144631 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein VITISV_024559 [Vitis vinifera] |
| 19 |
Hb_002999_070 |
0.1450525812 |
- |
- |
glucose-methanol-choline (gmc) oxidoreductase, putative [Ricinus communis] |
| 20 |
Hb_001014_020 |
0.1468656914 |
- |
- |
gene GA family protein [Populus trichocarpa] |