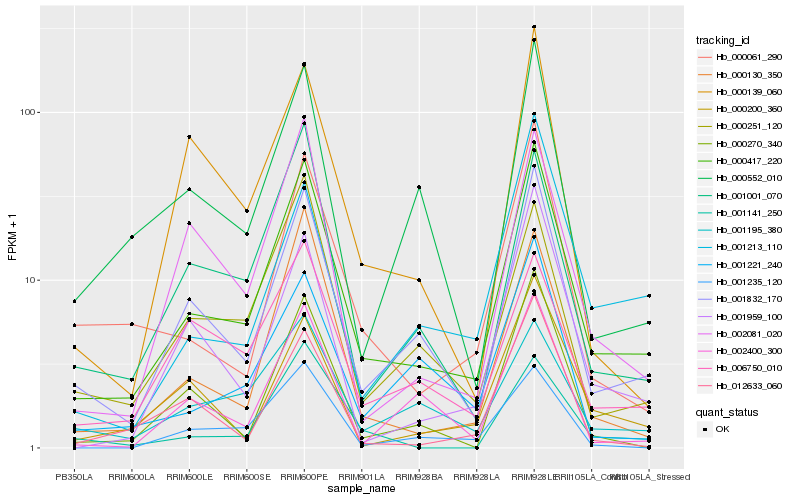
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000417_220 |
0.0 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 8 [UDP-forming] [Cucumis melo] |
| 2 |
Hb_001832_170 |
0.1048709868 |
- |
- |
PREDICTED: beta-galactosidase-like [Jatropha curcas] |
| 3 |
Hb_000061_290 |
0.1538430566 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000130_350 |
0.1561503224 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 5 |
Hb_001959_100 |
0.1590665428 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 6 |
Hb_000251_120 |
0.1628838506 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 7 |
Hb_001001_070 |
0.1634267378 |
- |
- |
PREDICTED: uncharacterized protein LOC105633663 [Jatropha curcas] |
| 8 |
Hb_002081_020 |
0.1642838267 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 9 |
Hb_000552_010 |
0.1681367529 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 2 [Jatropha curcas] |
| 10 |
Hb_000270_340 |
0.1686561048 |
- |
- |
hypothetical protein JCGZ_11546 [Jatropha curcas] |
| 11 |
Hb_000200_360 |
0.1693427804 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 12 |
Hb_001221_240 |
0.1709581517 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002400_300 |
0.1730880571 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 14 |
Hb_001141_250 |
0.1741370635 |
- |
- |
hypothetical protein glysoja_047929, partial [Glycine soja] |
| 15 |
Hb_001195_380 |
0.174447272 |
- |
- |
PREDICTED: uncharacterized protein LOC105633781 [Jatropha curcas] |
| 16 |
Hb_012633_060 |
0.1749317113 |
- |
- |
laccase, putative [Ricinus communis] |
| 17 |
Hb_000139_060 |
0.1760297808 |
- |
- |
Photosystem I reaction center subunit II, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_006750_010 |
0.177344476 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 19 |
Hb_001235_120 |
0.1774511516 |
- |
- |
hypothetical protein JCGZ_03395 [Jatropha curcas] |
| 20 |
Hb_001213_110 |
0.1776332162 |
- |
- |
SOUL heme-binding family protein [Populus trichocarpa] |