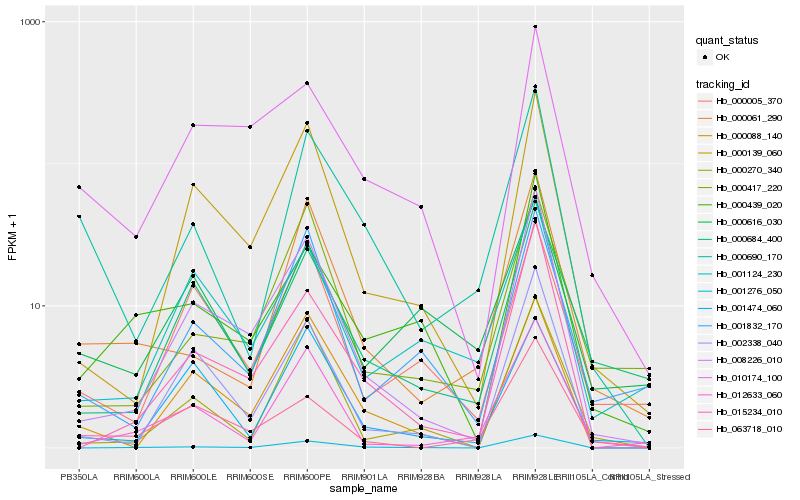
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000690_170 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000061_290 |
0.1657923812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000088_140 |
0.2116607255 |
- |
- |
- |
| 4 |
Hb_012633_060 |
0.2199340103 |
- |
- |
laccase, putative [Ricinus communis] |
| 5 |
Hb_008226_010 |
0.227062582 |
- |
- |
PREDICTED: uncharacterized protein LOC105649179 [Jatropha curcas] |
| 6 |
Hb_001276_050 |
0.2279880203 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance-like protein 1O [Medicago truncatula] |
| 7 |
Hb_001832_170 |
0.2324354836 |
- |
- |
PREDICTED: beta-galactosidase-like [Jatropha curcas] |
| 8 |
Hb_063718_010 |
0.2330842817 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000270_340 |
0.2338145647 |
- |
- |
hypothetical protein JCGZ_11546 [Jatropha curcas] |
| 10 |
Hb_000684_400 |
0.2355876429 |
- |
- |
PREDICTED: uncharacterized protein LOC105642159 [Jatropha curcas] |
| 11 |
Hb_000616_030 |
0.2362438015 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 12 |
Hb_000139_060 |
0.2362586571 |
- |
- |
Photosystem I reaction center subunit II, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_000417_220 |
0.2400419978 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 8 [UDP-forming] [Cucumis melo] |
| 14 |
Hb_015234_010 |
0.2412548706 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 4B [Jatropha curcas] |
| 15 |
Hb_002338_040 |
0.2422433774 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001474_060 |
0.2425688104 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 16 [Jatropha curcas] |
| 17 |
Hb_010174_100 |
0.2441331147 |
- |
- |
PREDICTED: photosystem II core complex proteins psbY, chloroplastic-like [Populus euphratica] |
| 18 |
Hb_000439_020 |
0.247528002 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3-like [Jatropha curcas] |
| 19 |
Hb_000005_370 |
0.2483967697 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105634024 [Jatropha curcas] |
| 20 |
Hb_001124_230 |
0.2504125401 |
- |
- |
PREDICTED: divinyl chlorophyllide a 8-vinyl-reductase, chloroplastic [Jatropha curcas] |