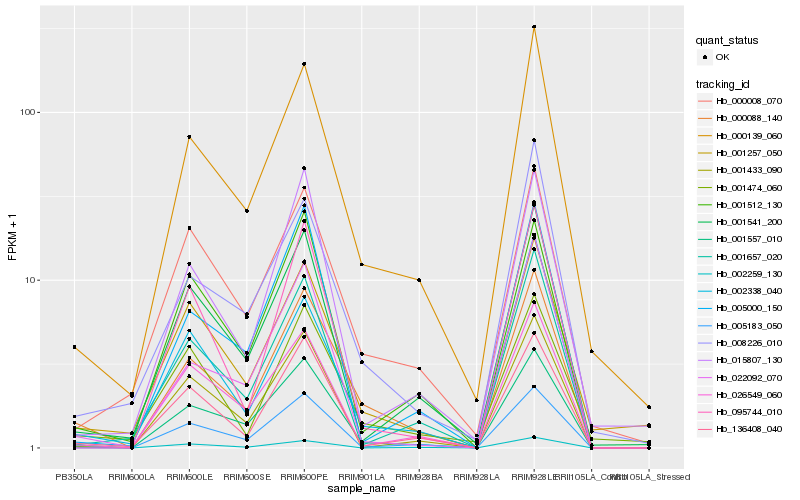
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000088_140 |
0.0 |
- |
- |
- |
| 2 |
Hb_001557_010 |
0.1054150885 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 3 |
Hb_000139_060 |
0.1096437389 |
- |
- |
Photosystem I reaction center subunit II, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_136408_040 |
0.1419503523 |
- |
- |
PREDICTED: protein NDR1 [Jatropha curcas] |
| 5 |
Hb_001257_050 |
0.1459843721 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: protein YLS9 [Jatropha curcas] |
| 6 |
Hb_005183_050 |
0.1487884704 |
- |
- |
hypothetical protein RCOM_1590900 [Ricinus communis] |
| 7 |
Hb_000008_070 |
0.1492041668 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_001541_200 |
0.1514086955 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 9 |
Hb_008226_010 |
0.1524837864 |
- |
- |
PREDICTED: uncharacterized protein LOC105649179 [Jatropha curcas] |
| 10 |
Hb_002338_040 |
0.1532808808 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_022092_070 |
0.1540557543 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.5 [Jatropha curcas] |
| 12 |
Hb_001474_060 |
0.1543130172 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 16 [Jatropha curcas] |
| 13 |
Hb_026549_060 |
0.1546601639 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 14 |
Hb_001657_020 |
0.1552535962 |
- |
- |
heat shock family protein [Populus trichocarpa] |
| 15 |
Hb_001433_090 |
0.1566655133 |
- |
- |
Pectinesterase PPE8B precursor, putative [Ricinus communis] |
| 16 |
Hb_002259_130 |
0.1588653915 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_005000_150 |
0.159188095 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_095744_010 |
0.1601018809 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 [Fragaria vesca subsp. vesca] |
| 19 |
Hb_001512_130 |
0.1607132538 |
- |
- |
PREDICTED: uncharacterized protein LOC105633633 [Jatropha curcas] |
| 20 |
Hb_015807_130 |
0.1615243381 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |