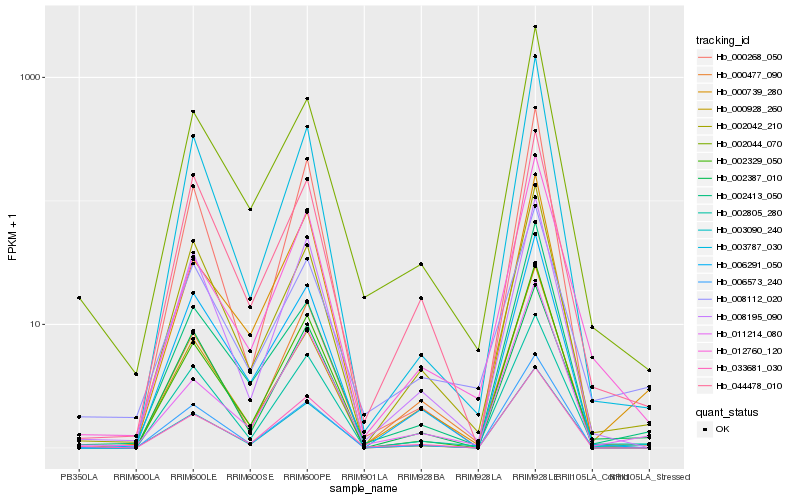
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003090_240 |
0.0 |
- |
- |
hypothetical protein JCGZ_25095 [Jatropha curcas] |
| 2 |
Hb_002805_280 |
0.0734792433 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 3 |
Hb_002387_010 |
0.0840511788 |
- |
- |
PREDICTED: protein STAY-GREEN LIKE, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002042_210 |
0.0951772349 |
- |
- |
PREDICTED: UPF0603 protein At1g54780, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000739_280 |
0.0983787608 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_008195_090 |
0.1016235147 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_006291_050 |
0.1056530726 |
- |
- |
PREDICTED: polyribonucleotide nucleotidyltransferase 2, mitochondrial isoform X1 [Jatropha curcas] |
| 8 |
Hb_006573_240 |
0.106205683 |
- |
- |
PREDICTED: anthocyanidin 5,3-O-glucosyltransferase-like [Jatropha curcas] |
| 9 |
Hb_002329_050 |
0.1069982923 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003787_030 |
0.1089127118 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000477_090 |
0.1109533294 |
- |
- |
PREDICTED: uncharacterized protein LOC105639302 [Jatropha curcas] |
| 12 |
Hb_033681_030 |
0.1117129714 |
desease resistance |
Gene Name: NB-ARC |
Leucine-rich repeat containing protein [Theobroma cacao] |
| 13 |
Hb_002044_070 |
0.1125797669 |
- |
- |
Chlorophyll a-b binding protein 3, chloroplastic [Theobroma cacao] |
| 14 |
Hb_011214_080 |
0.1129496792 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 15 |
Hb_000268_050 |
0.1131645071 |
- |
- |
PREDICTED: tetrapyrrole-binding protein, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002413_050 |
0.1156833842 |
- |
- |
hypothetical protein POPTR_0018s06150g [Populus trichocarpa] |
| 17 |
Hb_012760_120 |
0.1157384656 |
- |
- |
PREDICTED: peroxidase 64-like [Jatropha curcas] |
| 18 |
Hb_044478_010 |
0.1160242438 |
- |
- |
plastoquinol-plastocyanin reductase, putative [Ricinus communis] |
| 19 |
Hb_000928_260 |
0.1165159842 |
- |
- |
hypothetical protein POPTR_0019s06080g [Populus trichocarpa] |
| 20 |
Hb_008112_020 |
0.1196725718 |
- |
- |
PREDICTED: magnesium transporter MRS2-11, chloroplastic [Jatropha curcas] |