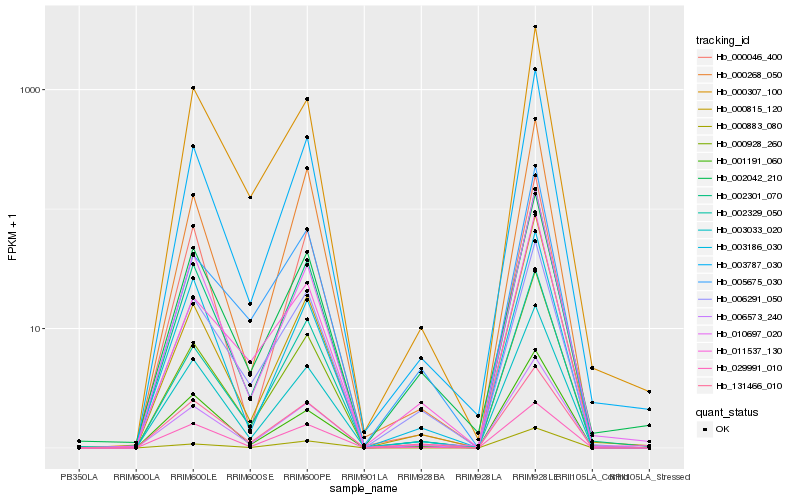
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006573_240 |
0.0 |
- |
- |
PREDICTED: anthocyanidin 5,3-O-glucosyltransferase-like [Jatropha curcas] |
| 2 |
Hb_003033_020 |
0.0434890719 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9-like [Jatropha curcas] |
| 3 |
Hb_003787_030 |
0.0500712537 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000928_260 |
0.0520764195 |
- |
- |
hypothetical protein POPTR_0019s06080g [Populus trichocarpa] |
| 5 |
Hb_002301_070 |
0.0594579635 |
- |
- |
PREDICTED: triose phosphate/phosphate translocator TPT, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_002329_050 |
0.0627702371 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000307_100 |
0.0705340063 |
- |
- |
Plastocyanin A, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_006291_050 |
0.0745301702 |
- |
- |
PREDICTED: polyribonucleotide nucleotidyltransferase 2, mitochondrial isoform X1 [Jatropha curcas] |
| 9 |
Hb_010697_020 |
0.0752018481 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_131466_010 |
0.0763103014 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 11 |
Hb_000268_050 |
0.077547387 |
- |
- |
PREDICTED: tetrapyrrole-binding protein, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002042_210 |
0.0815723033 |
- |
- |
PREDICTED: UPF0603 protein At1g54780, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001191_060 |
0.081734021 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Populus euphratica] |
| 14 |
Hb_011537_130 |
0.0857525764 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_000883_080 |
0.0869127587 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_003186_030 |
0.087123017 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0012s05720g [Populus trichocarpa] |
| 17 |
Hb_029991_010 |
0.0872264671 |
- |
- |
hypothetical protein PRUPE_ppa001615mg [Prunus persica] |
| 18 |
Hb_005675_030 |
0.0880310842 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 6 [Jatropha curcas] |
| 19 |
Hb_000815_120 |
0.0897953378 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 20 |
Hb_000046_400 |
0.090667741 |
- |
- |
conserved hypothetical protein [Ricinus communis] |