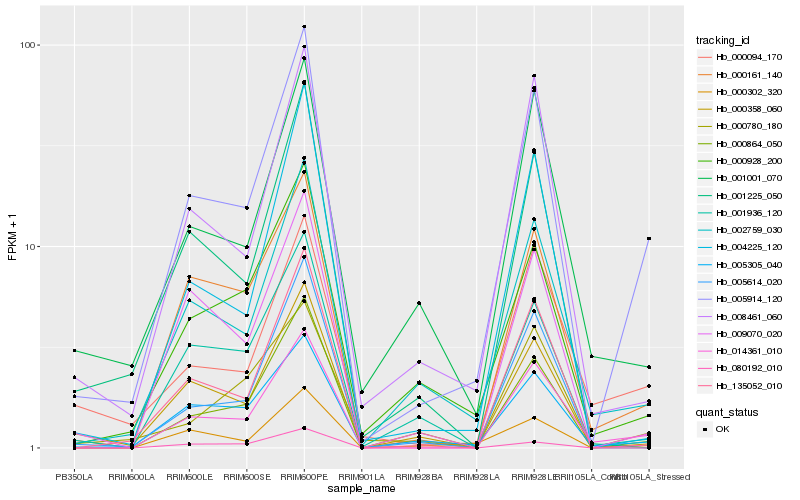
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005914_120 |
0.0 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 2B [Jatropha curcas] |
| 2 |
Hb_002759_030 |
0.1352399603 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 [Jatropha curcas] |
| 3 |
Hb_005305_040 |
0.1430335102 |
transcription factor |
TF Family: C3H |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_000161_140 |
0.1518826778 |
- |
- |
PREDICTED: WEB family protein At3g51220 [Jatropha curcas] |
| 5 |
Hb_014361_010 |
0.1554774958 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 6 |
Hb_008461_060 |
0.1581024798 |
- |
- |
hypothetical protein JCGZ_17379 [Jatropha curcas] |
| 7 |
Hb_000864_050 |
0.1582511936 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_000302_320 |
0.1584566019 |
- |
- |
hypothetical protein JCGZ_26057 [Jatropha curcas] |
| 9 |
Hb_000358_060 |
0.1589641638 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 10 |
Hb_000094_170 |
0.1608622443 |
- |
- |
PREDICTED: uncharacterized protein LOC105633781 [Jatropha curcas] |
| 11 |
Hb_080192_010 |
0.1610229574 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 12 |
Hb_000928_200 |
0.1630992679 |
- |
- |
PREDICTED: RING-H2 finger protein ATL52-like [Jatropha curcas] |
| 13 |
Hb_004225_120 |
0.1635163271 |
- |
- |
PREDICTED: uncharacterized protein LOC105628059 [Jatropha curcas] |
| 14 |
Hb_009070_020 |
0.166166063 |
- |
- |
PREDICTED: uncharacterized protein LOC102612959 [Citrus sinensis] |
| 15 |
Hb_135052_010 |
0.1684660705 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 16 |
Hb_005614_020 |
0.1720882158 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001936_120 |
0.1734617562 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001225_050 |
0.1736922361 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 19 |
Hb_001001_070 |
0.1738281766 |
- |
- |
PREDICTED: uncharacterized protein LOC105633663 [Jatropha curcas] |
| 20 |
Hb_000780_180 |
0.1749658404 |
- |
- |
PREDICTED: uncharacterized protein LOC105641547 [Jatropha curcas] |