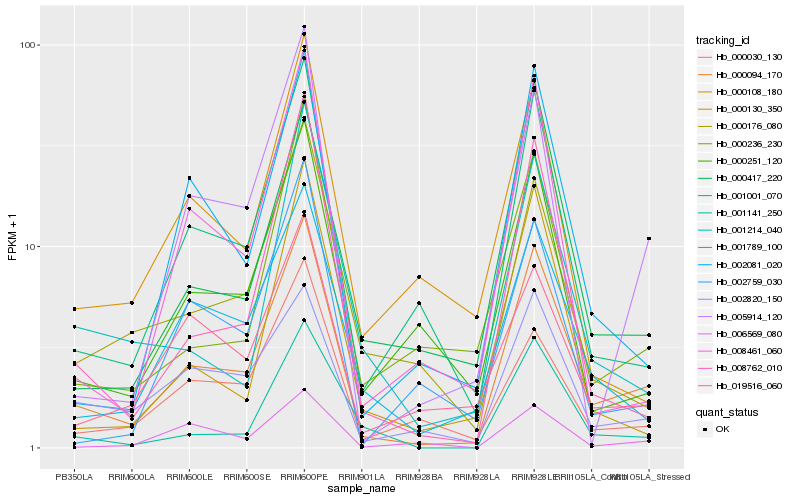
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000094_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633781 [Jatropha curcas] |
| 2 |
Hb_001001_070 |
0.1372564009 |
- |
- |
PREDICTED: uncharacterized protein LOC105633663 [Jatropha curcas] |
| 3 |
Hb_008762_010 |
0.1543556558 |
- |
- |
dopamine beta-monooxygenase, putative [Ricinus communis] |
| 4 |
Hb_001789_100 |
0.1553256151 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001141_250 |
0.1565057034 |
- |
- |
hypothetical protein glysoja_047929, partial [Glycine soja] |
| 6 |
Hb_002081_020 |
0.1597343861 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 7 |
Hb_005914_120 |
0.1608622443 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 2B [Jatropha curcas] |
| 8 |
Hb_002820_150 |
0.1611953201 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_000251_120 |
0.1644700776 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 10 |
Hb_008461_060 |
0.1683243708 |
- |
- |
hypothetical protein JCGZ_17379 [Jatropha curcas] |
| 11 |
Hb_000176_080 |
0.1713112964 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g30520 [Jatropha curcas] |
| 12 |
Hb_000236_230 |
0.175256392 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At4g11680 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000030_130 |
0.1761064377 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_019516_060 |
0.1776762654 |
- |
- |
PREDICTED: uncharacterized protein LOC105634259 [Jatropha curcas] |
| 15 |
Hb_000417_220 |
0.179004026 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 8 [UDP-forming] [Cucumis melo] |
| 16 |
Hb_001214_040 |
0.1791458161 |
- |
- |
PREDICTED: protein trichome birefringence-like 3 [Jatropha curcas] |
| 17 |
Hb_000130_350 |
0.1822224523 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 18 |
Hb_002759_030 |
0.1854120749 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 [Jatropha curcas] |
| 19 |
Hb_000108_180 |
0.1856000781 |
- |
- |
PREDICTED: uncharacterized protein LOC105630021 [Jatropha curcas] |
| 20 |
Hb_006569_080 |
0.1874984927 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein VITISV_024559 [Vitis vinifera] |