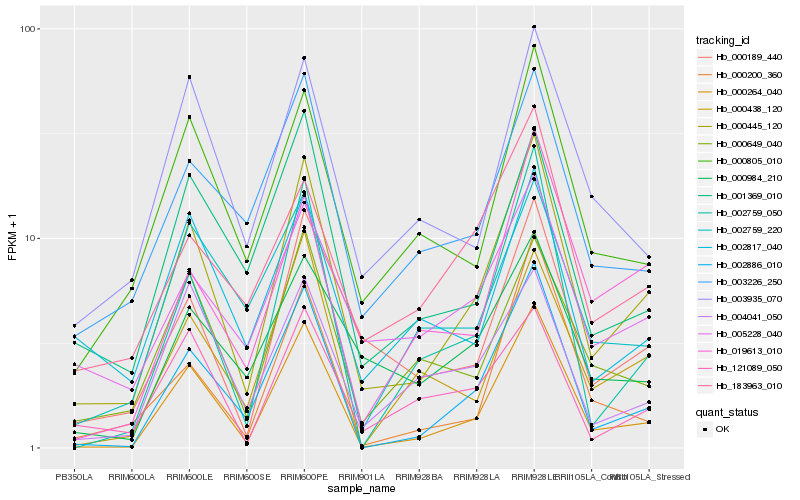
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000445_120 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_000264_040 |
0.1114219274 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 3 |
Hb_002886_010 |
0.1230764356 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000189_440 |
0.139958075 |
- |
- |
hypothetical protein JCGZ_23470 [Jatropha curcas] |
| 5 |
Hb_000805_010 |
0.1558355597 |
- |
- |
PREDICTED: uncharacterized protein LOC105629931 [Jatropha curcas] |
| 6 |
Hb_000649_040 |
0.1585855779 |
- |
- |
PREDICTED: dynamin-related protein 1C [Jatropha curcas] |
| 7 |
Hb_002759_050 |
0.1614418901 |
- |
- |
PREDICTED: protein ALTERED XYLOGLUCAN 4 [Jatropha curcas] |
| 8 |
Hb_003226_250 |
0.1627935341 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 9 |
Hb_001369_010 |
0.1640858488 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 10 |
Hb_003935_070 |
0.1677546031 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005228_040 |
0.1682449926 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 12 |
Hb_121089_050 |
0.1689911329 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000438_120 |
0.1740524369 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 14 |
Hb_002817_040 |
0.1761032277 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |
| 15 |
Hb_004041_050 |
0.1768915543 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 16 |
Hb_000984_210 |
0.1772953937 |
- |
- |
PREDICTED: uncharacterized protein LOC104451821 [Eucalyptus grandis] |
| 17 |
Hb_019613_010 |
0.1777968144 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 18 |
Hb_002759_220 |
0.1804681364 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 19 |
Hb_183963_010 |
0.1806830211 |
- |
- |
core region of GTP cyclohydrolase I family protein [Populus trichocarpa] |
| 20 |
Hb_000200_360 |
0.182671684 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |