| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
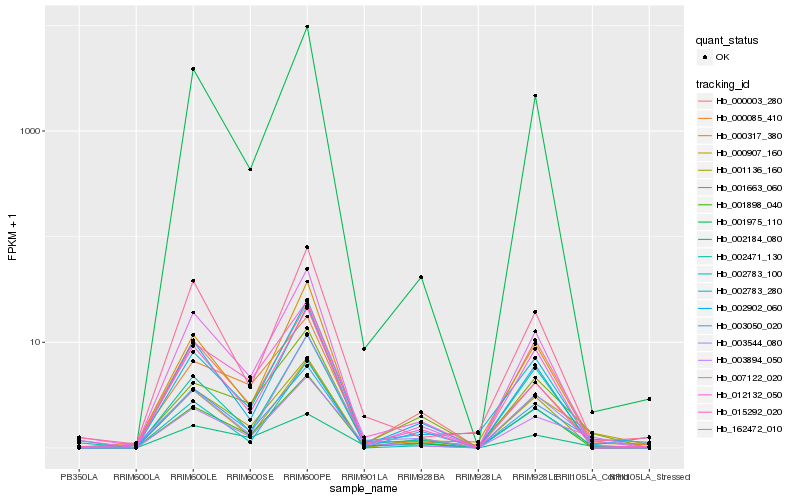
Hb_003894_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_14524 [Jatropha curcas] |
| 2 |
Hb_003544_080 |
0.1342731466 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 3 |
Hb_000907_160 |
0.1378425547 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 4 |
Hb_002783_100 |
0.1543664095 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 5 |
Hb_001898_040 |
0.1584738755 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 6 |
Hb_001663_060 |
0.1628617825 |
- |
- |
ribonuclease t2, putative [Ricinus communis] |
| 7 |
Hb_001136_160 |
0.1633073882 |
- |
- |
catalytic, putative [Ricinus communis] |
| 8 |
Hb_000317_380 |
0.1679378186 |
- |
- |
Peroxidase 47 precursor, putative [Ricinus communis] |
| 9 |
Hb_162472_010 |
0.1681404732 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_015292_020 |
0.1682020305 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_003050_020 |
0.1685764003 |
- |
- |
glycosyl hydrolase family 17 family protein [Populus trichocarpa] |
| 12 |
Hb_007122_020 |
0.1687478166 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 13 |
Hb_001975_110 |
0.1711229463 |
- |
- |
- |
| 14 |
Hb_002783_280 |
0.1729256989 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 15 |
Hb_000003_280 |
0.1730822075 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_012132_050 |
0.1735338564 |
- |
- |
PREDICTED: aspartic proteinase PCS1 [Jatropha curcas] |
| 17 |
Hb_002184_080 |
0.174851824 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 18 |
Hb_000085_410 |
0.1751424954 |
- |
- |
hypothetical protein POPTR_0011s09040g [Populus trichocarpa] |
| 19 |
Hb_002471_130 |
0.1757889088 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 3 [Jatropha curcas] |
| 20 |
Hb_002902_060 |
0.1773214765 |
- |
- |
PREDICTED: uncharacterized protein LOC105639824 [Jatropha curcas] |