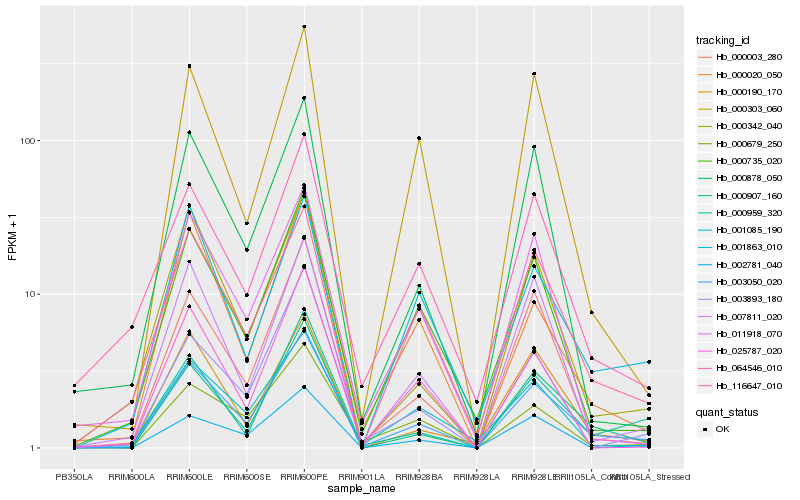
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003050_020 |
0.0 |
- |
- |
glycosyl hydrolase family 17 family protein [Populus trichocarpa] |
| 2 |
Hb_000907_160 |
0.1066374205 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 3 |
Hb_000735_020 |
0.117528713 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 4 |
Hb_116647_010 |
0.1201152837 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000003_280 |
0.1213552318 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000959_320 |
0.1255283835 |
- |
- |
PREDICTED: uncharacterized protein LOC105778325 [Gossypium raimondii] |
| 7 |
Hb_007811_020 |
0.1269489049 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |
| 8 |
Hb_000190_170 |
0.1295129917 |
- |
- |
PREDICTED: chlorophyllase-1-like [Jatropha curcas] |
| 9 |
Hb_000342_040 |
0.1297457561 |
- |
- |
pollen specific protein sf21, putative [Ricinus communis] |
| 10 |
Hb_002781_040 |
0.1361661457 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001085_190 |
0.136185618 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 12 |
Hb_064546_010 |
0.1367369007 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 13 |
Hb_000878_050 |
0.138165006 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 14 |
Hb_003893_180 |
0.1381819673 |
- |
- |
PREDICTED: uncharacterized protein LOC105648771 [Jatropha curcas] |
| 15 |
Hb_011918_070 |
0.1402686738 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 16 |
Hb_001863_010 |
0.1407414194 |
- |
- |
PREDICTED: classical arabinogalactan protein 4-like [Jatropha curcas] |
| 17 |
Hb_000303_060 |
0.1407639673 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 18 |
Hb_000020_050 |
0.1411778341 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |
| 19 |
Hb_000679_250 |
0.1416324848 |
- |
- |
PREDICTED: uncharacterized protein LOC105637632 isoform X1 [Jatropha curcas] |
| 20 |
Hb_025787_020 |
0.142885291 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |