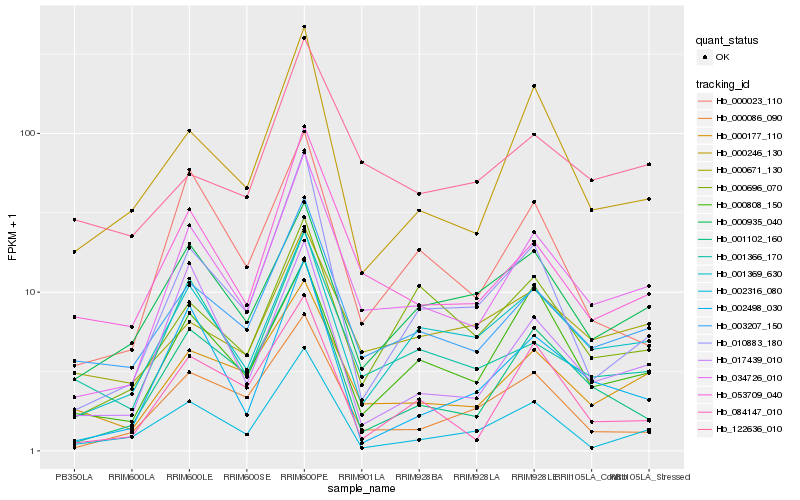
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_034726_010 |
0.0 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X2 [Jatropha curcas] |
| 2 |
Hb_003207_150 |
0.1145018738 |
- |
- |
PREDICTED: SPX domain-containing membrane protein At4g22990-like [Jatropha curcas] |
| 3 |
Hb_000177_110 |
0.1203577735 |
- |
- |
PREDICTED: uncharacterized protein LOC105642063 [Jatropha curcas] |
| 4 |
Hb_053709_040 |
0.1254937853 |
- |
- |
putative actin-97 protein [Linum usitatissimum] |
| 5 |
Hb_001369_630 |
0.1318118059 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 6 |
Hb_001102_160 |
0.1434350251 |
- |
- |
PREDICTED: galacturonokinase [Jatropha curcas] |
| 7 |
Hb_001366_170 |
0.1482132311 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 8 |
Hb_000808_150 |
0.1500643336 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 9 |
Hb_000246_130 |
0.1507212603 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 10 |
Hb_000086_090 |
0.1538099696 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |
| 11 |
Hb_010883_180 |
0.1574629528 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2-like [Jatropha curcas] |
| 12 |
Hb_000023_110 |
0.1587557498 |
- |
- |
PREDICTED: probable methyltransferase PMT21 [Jatropha curcas] |
| 13 |
Hb_084147_010 |
0.1616683034 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 14 |
Hb_002498_030 |
0.1624202263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000935_040 |
0.1649783657 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 16 |
Hb_017439_010 |
0.1650314226 |
- |
- |
putative actin-97 protein [Linum usitatissimum] |
| 17 |
Hb_000671_130 |
0.1731717106 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000696_070 |
0.1737887067 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 [Jatropha curcas] |
| 19 |
Hb_122636_010 |
0.1743078197 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 20 |
Hb_002316_080 |
0.1745970861 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46-like [Populus euphratica] |