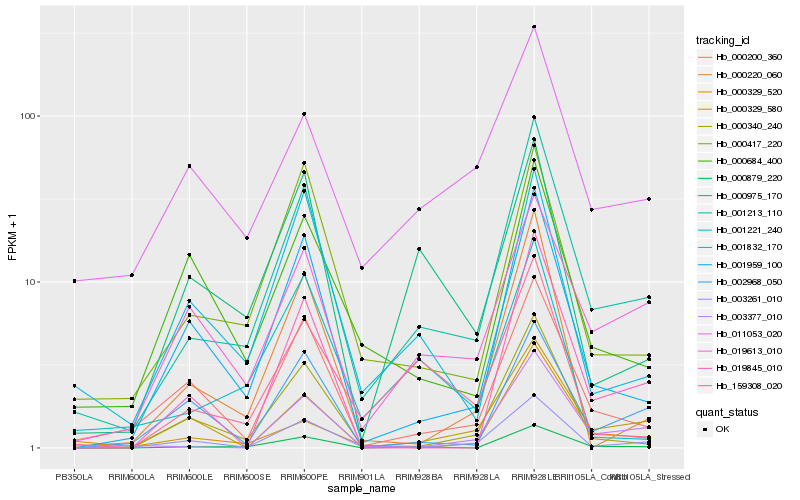
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001213_110 |
0.0 |
- |
- |
SOUL heme-binding family protein [Populus trichocarpa] |
| 2 |
Hb_000879_220 |
0.13876421 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 3 |
Hb_003261_010 |
0.1603273998 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 4 |
Hb_001959_100 |
0.1712498116 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 5 |
Hb_003377_010 |
0.1729274784 |
- |
- |
hypothetical protein POPTR_0016s06140g, partial [Populus trichocarpa] |
| 6 |
Hb_000417_220 |
0.1776332162 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 8 [UDP-forming] [Cucumis melo] |
| 7 |
Hb_000200_360 |
0.1781916794 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 8 |
Hb_019613_010 |
0.1786435254 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_000340_240 |
0.1817088861 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000329_580 |
0.1911952584 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_011053_020 |
0.1956248303 |
- |
- |
lipoic acid synthetase, putative [Ricinus communis] |
| 12 |
Hb_000220_060 |
0.1974618921 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 15 [Jatropha curcas] |
| 13 |
Hb_001832_170 |
0.1983664037 |
- |
- |
PREDICTED: beta-galactosidase-like [Jatropha curcas] |
| 14 |
Hb_159308_020 |
0.200058878 |
- |
- |
hypothetical protein B456_002G027900 [Gossypium raimondii] |
| 15 |
Hb_019845_010 |
0.2031901409 |
- |
- |
protease inhibitor protein [Hevea brasiliensis] |
| 16 |
Hb_002968_050 |
0.210825389 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 17 |
Hb_000329_520 |
0.210891438 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 18 |
Hb_001221_240 |
0.2119540927 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000975_170 |
0.2140221125 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 6 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000684_400 |
0.2176196237 |
- |
- |
PREDICTED: uncharacterized protein LOC105642159 [Jatropha curcas] |