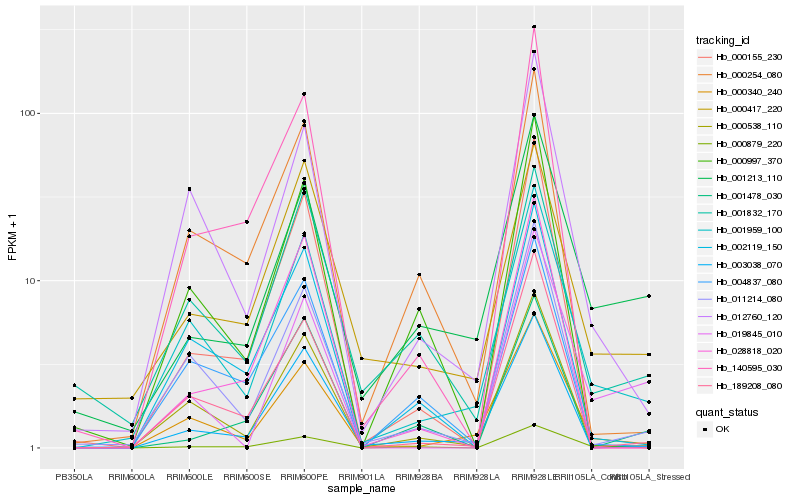
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000879_220 |
0.0 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 2 |
Hb_001213_110 |
0.13876421 |
- |
- |
SOUL heme-binding family protein [Populus trichocarpa] |
| 3 |
Hb_001959_100 |
0.1577697006 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 4 |
Hb_019845_010 |
0.1697302438 |
- |
- |
protease inhibitor protein [Hevea brasiliensis] |
| 5 |
Hb_001478_030 |
0.1705652731 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002119_150 |
0.1836048074 |
- |
- |
hypothetical protein POPTR_0008s13870g [Populus trichocarpa] |
| 7 |
Hb_000340_240 |
0.1860731646 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000538_110 |
0.1875505437 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 9 |
Hb_000155_230 |
0.1905990411 |
- |
- |
rac gtpase, putative [Ricinus communis] |
| 10 |
Hb_189208_080 |
0.1907457218 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 11-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000417_220 |
0.1908005253 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 8 [UDP-forming] [Cucumis melo] |
| 12 |
Hb_000254_080 |
0.1926798814 |
- |
- |
PREDICTED: uncharacterized protein LOC105635000 [Jatropha curcas] |
| 13 |
Hb_011214_080 |
0.1944551547 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 14 |
Hb_003038_070 |
0.195463106 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 15 |
Hb_012760_120 |
0.1956281779 |
- |
- |
PREDICTED: peroxidase 64-like [Jatropha curcas] |
| 16 |
Hb_000997_370 |
0.1969719059 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Jatropha curcas] |
| 17 |
Hb_028818_020 |
0.1978401486 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9-like [Jatropha curcas] |
| 18 |
Hb_004837_080 |
0.1997994977 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46 [Jatropha curcas] |
| 19 |
Hb_001832_170 |
0.2000114475 |
- |
- |
PREDICTED: beta-galactosidase-like [Jatropha curcas] |
| 20 |
Hb_140595_030 |
0.2000233382 |
- |
- |
- |