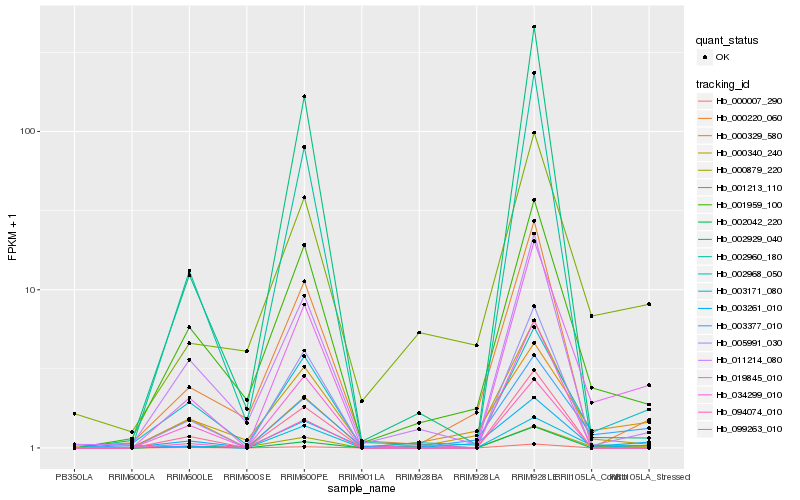
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019845_010 |
0.0 |
- |
- |
protease inhibitor protein [Hevea brasiliensis] |
| 2 |
Hb_000879_220 |
0.1697302438 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 3 |
Hb_002968_050 |
0.1861558293 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 4 |
Hb_003377_010 |
0.1864479265 |
- |
- |
hypothetical protein POPTR_0016s06140g, partial [Populus trichocarpa] |
| 5 |
Hb_003261_010 |
0.1889075745 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 6 |
Hb_001959_100 |
0.1904262295 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 7 |
Hb_000340_240 |
0.1981846655 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003171_080 |
0.2005595294 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001213_110 |
0.2031901409 |
- |
- |
SOUL heme-binding family protein [Populus trichocarpa] |
| 10 |
Hb_002960_180 |
0.2044189245 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 11 |
Hb_000220_060 |
0.2071601633 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 15 [Jatropha curcas] |
| 12 |
Hb_099263_010 |
0.2080812934 |
- |
- |
hypothetical protein AALP_AAs50029U000400 [Arabis alpina] |
| 13 |
Hb_094074_010 |
0.2090014136 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 14 |
Hb_005991_030 |
0.2090830861 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 15 |
Hb_002929_040 |
0.2097145279 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_000329_580 |
0.2107402562 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_011214_080 |
0.2129447471 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 18 |
Hb_034299_010 |
0.2129510908 |
- |
- |
F-box and wd40 domain protein, putative [Ricinus communis] |
| 19 |
Hb_002042_220 |
0.2148742031 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 20 |
Hb_000007_290 |
0.2151259776 |
- |
- |
hypothetical protein AALP_AAs43195U000200 [Arabis alpina] |