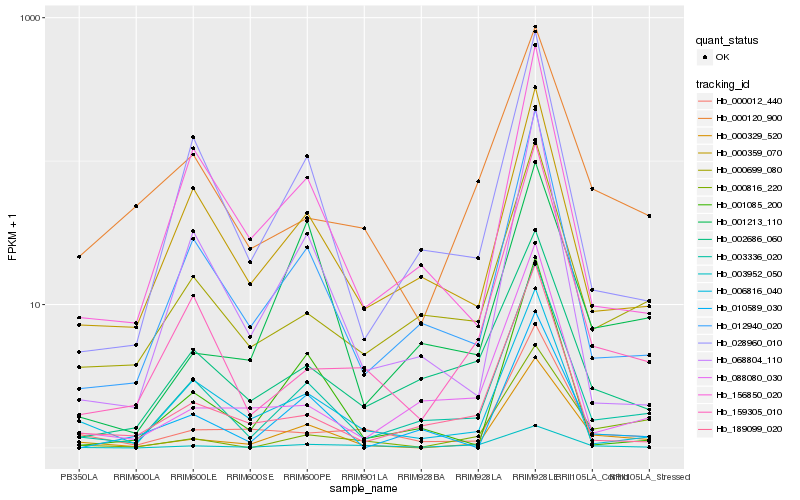
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_520 |
0.0 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 2 |
Hb_003336_020 |
0.1663012535 |
- |
- |
PREDICTED: probable sodium/metabolite cotransporter BASS3, chloroplastic [Vitis vinifera] |
| 3 |
Hb_000012_440 |
0.1919317609 |
- |
- |
PREDICTED: uncharacterized protein LOC104248335 [Nicotiana sylvestris] |
| 4 |
Hb_000816_220 |
0.1939621495 |
- |
- |
PREDICTED: CBL-interacting protein kinase 18-like [Jatropha curcas] |
| 5 |
Hb_012940_020 |
0.1990516762 |
- |
- |
zeaxanthin epoxidase, putative [Ricinus communis] |
| 6 |
Hb_000699_080 |
0.2107671074 |
- |
- |
PREDICTED: rubisco accumulation factor 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001213_110 |
0.210891438 |
- |
- |
SOUL heme-binding family protein [Populus trichocarpa] |
| 8 |
Hb_159305_010 |
0.2138178876 |
- |
- |
rpl2 [Jatropha curcas] |
| 9 |
Hb_003952_050 |
0.2182002402 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 10 |
Hb_006816_040 |
0.2196950721 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48910-like [Jatropha curcas] |
| 11 |
Hb_000120_900 |
0.2201728881 |
- |
- |
catalase [Hevea brasiliensis] |
| 12 |
Hb_000359_070 |
0.2235697466 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001085_200 |
0.2269679021 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 15 [Jatropha curcas] |
| 14 |
Hb_010589_030 |
0.2271447943 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK2 [Jatropha curcas] |
| 15 |
Hb_068804_110 |
0.2305639447 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_028960_010 |
0.2307592083 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 17 |
Hb_189099_020 |
0.2332709603 |
- |
- |
NADH dehydrogenase subunit 9 (mitochondrion) [Hevea brasiliensis] |
| 18 |
Hb_088080_030 |
0.2344242372 |
- |
- |
ribosomal protein S4 [Hevea brasiliensis] |
| 19 |
Hb_002686_060 |
0.2346506033 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_156850_020 |
0.234935253 |
- |
- |
ATP-dependent zinc metalloprotease FTSH 1, chloroplastic [Glycine soja] |