| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
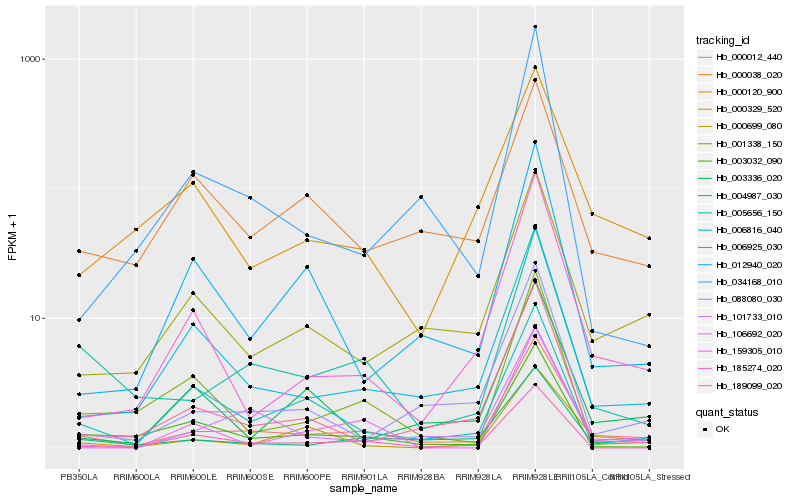
Hb_000012_440 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104248335 [Nicotiana sylvestris] |
| 2 |
Hb_004987_030 |
0.1355181912 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 3 |
Hb_005656_150 |
0.1448398606 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 4 |
Hb_000699_080 |
0.1573564627 |
- |
- |
PREDICTED: rubisco accumulation factor 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_006925_030 |
0.1580572824 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18840-like [Populus euphratica] |
| 6 |
Hb_088080_030 |
0.1769721702 |
- |
- |
ribosomal protein S4 [Hevea brasiliensis] |
| 7 |
Hb_003032_090 |
0.180315433 |
- |
- |
hypothetical protein POPTR_0018s06450g [Populus trichocarpa] |
| 8 |
Hb_189099_020 |
0.1818673519 |
- |
- |
NADH dehydrogenase subunit 9 (mitochondrion) [Hevea brasiliensis] |
| 9 |
Hb_159305_010 |
0.1829313089 |
- |
- |
rpl2 [Jatropha curcas] |
| 10 |
Hb_006816_040 |
0.1881119445 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48910-like [Jatropha curcas] |
| 11 |
Hb_000120_900 |
0.1887337108 |
- |
- |
catalase [Hevea brasiliensis] |
| 12 |
Hb_101733_010 |
0.189924218 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000329_520 |
0.1919317609 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 14 |
Hb_000038_020 |
0.1943600597 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_003336_020 |
0.1968910466 |
- |
- |
PREDICTED: probable sodium/metabolite cotransporter BASS3, chloroplastic [Vitis vinifera] |
| 16 |
Hb_012940_020 |
0.19698379 |
- |
- |
zeaxanthin epoxidase, putative [Ricinus communis] |
| 17 |
Hb_106692_020 |
0.1974504542 |
- |
- |
NADH dehydrogenase subunit F, partial (chloroplast) [Hevea sp. Gillespie 4272] |
| 18 |
Hb_034168_010 |
0.197605381 |
- |
- |
hypothetical protein [Azospirillum sp. CAG:260] |
| 19 |
Hb_185274_020 |
0.1993053067 |
- |
- |
Ycf2 (chloroplast) [Andrographis paniculata] |
| 20 |
Hb_001338_150 |
0.2007164275 |
- |
- |
hypothetical chloroplast RF1 [Dillenia indica] |