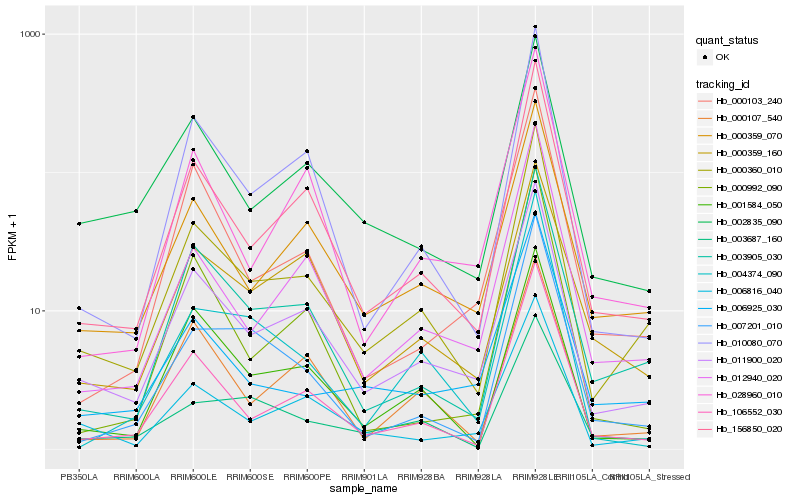
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000360_010 |
0.0 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_011900_020 |
0.0831725467 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g16390, chloroplastic [Jatropha curcas] |
| 3 |
Hb_156850_020 |
0.0918246704 |
- |
- |
ATP-dependent zinc metalloprotease FTSH 1, chloroplastic [Glycine soja] |
| 4 |
Hb_003905_030 |
0.1009738868 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 5 |
Hb_106552_030 |
0.1036538984 |
- |
- |
hypothetical protein POPTR_0006s23180g [Populus trichocarpa] |
| 6 |
Hb_000359_070 |
0.1082884606 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_010080_070 |
0.1178547281 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH, chloroplastic [Jatropha curcas] |
| 8 |
Hb_006816_040 |
0.122183016 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48910-like [Jatropha curcas] |
| 9 |
Hb_012940_020 |
0.1255220997 |
- |
- |
zeaxanthin epoxidase, putative [Ricinus communis] |
| 10 |
Hb_001584_050 |
0.1280153714 |
- |
- |
PREDICTED: NHL repeat-containing protein 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002835_090 |
0.1394315673 |
- |
- |
RecName: Full=Elongation factor Tu, chloroplastic; Short=EF-Tu; Flags: Precursor [Glycine max] |
| 12 |
Hb_006925_030 |
0.1400253042 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18840-like [Populus euphratica] |
| 13 |
Hb_000107_540 |
0.1404192428 |
- |
- |
PREDICTED: uncharacterized protein LOC105635335 [Jatropha curcas] |
| 14 |
Hb_028960_010 |
0.1447040811 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 15 |
Hb_003687_160 |
0.1453652358 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 16 |
Hb_007201_010 |
0.1456825068 |
- |
- |
PREDICTED: uncharacterized protein LOC105631226 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000103_240 |
0.1491886519 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 1 [Jatropha curcas] |
| 18 |
Hb_000359_160 |
0.1537929811 |
- |
- |
PREDICTED: protein CAJ1 [Jatropha curcas] |
| 19 |
Hb_004374_090 |
0.1565888445 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |
| 20 |
Hb_000992_090 |
0.1581341117 |
transcription factor |
TF Family: Orphans |
CIL, putative [Ricinus communis] |