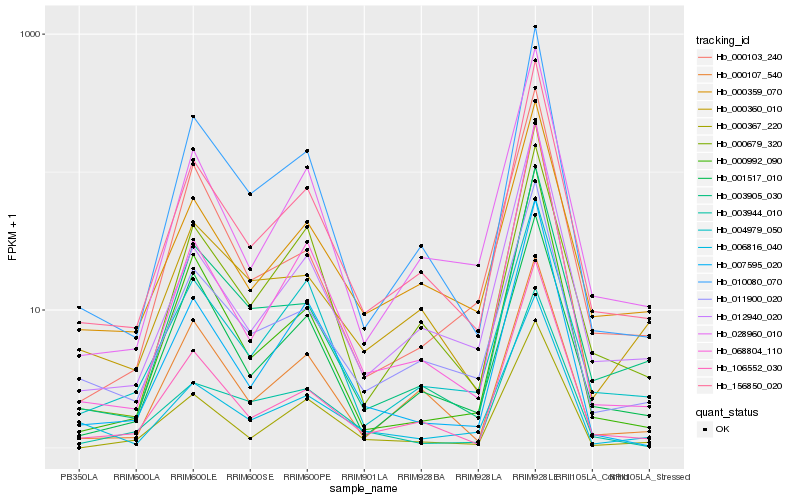
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_156850_020 |
0.0 |
- |
- |
ATP-dependent zinc metalloprotease FTSH 1, chloroplastic [Glycine soja] |
| 2 |
Hb_106552_030 |
0.0622294204 |
- |
- |
hypothetical protein POPTR_0006s23180g [Populus trichocarpa] |
| 3 |
Hb_010080_070 |
0.0700566903 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH, chloroplastic [Jatropha curcas] |
| 4 |
Hb_028960_010 |
0.0730146589 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 5 |
Hb_012940_020 |
0.0744382089 |
- |
- |
zeaxanthin epoxidase, putative [Ricinus communis] |
| 6 |
Hb_011900_020 |
0.0752425037 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g16390, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000359_070 |
0.0812588299 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000360_010 |
0.0918246704 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_068804_110 |
0.0930026321 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial [Jatropha curcas] |
| 10 |
Hb_003905_030 |
0.1010626505 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 11 |
Hb_000367_220 |
0.1040036744 |
- |
- |
hypothetical protein RCOM_1470580 [Ricinus communis] |
| 12 |
Hb_007595_020 |
0.1047888355 |
- |
- |
maturase K [Hevea brasiliensis] |
| 13 |
Hb_000992_090 |
0.1070789735 |
transcription factor |
TF Family: Orphans |
CIL, putative [Ricinus communis] |
| 14 |
Hb_006816_040 |
0.1073827945 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48910-like [Jatropha curcas] |
| 15 |
Hb_000107_540 |
0.1085572203 |
- |
- |
PREDICTED: uncharacterized protein LOC105635335 [Jatropha curcas] |
| 16 |
Hb_004979_050 |
0.1152287164 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 17 |
Hb_000679_320 |
0.118318143 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105136146 [Populus euphratica] |
| 18 |
Hb_001517_010 |
0.118458447 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003944_010 |
0.1186491402 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 20 |
Hb_000103_240 |
0.1198292932 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 1 [Jatropha curcas] |