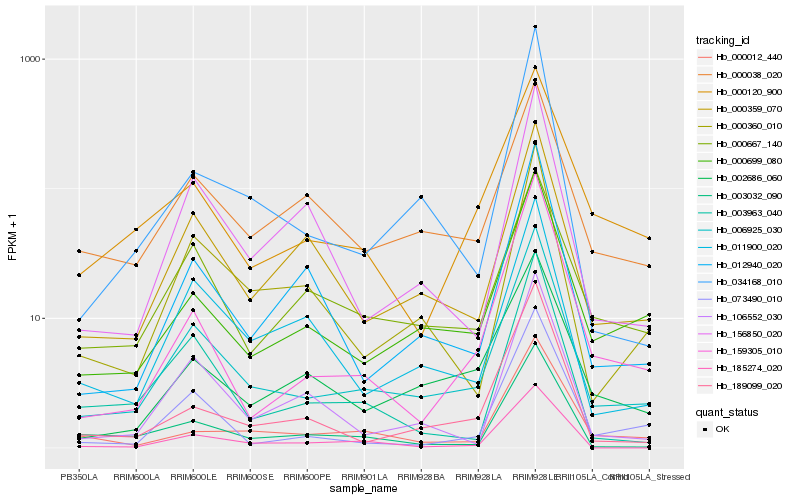
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006925_030 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18840-like [Populus euphratica] |
| 2 |
Hb_000699_080 |
0.1234856873 |
- |
- |
PREDICTED: rubisco accumulation factor 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000359_070 |
0.1379764657 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000360_010 |
0.1400253042 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_011900_020 |
0.1448342682 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g16390, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002686_060 |
0.1472245032 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_073490_010 |
0.1484773502 |
- |
- |
NAD(P)-linked oxidoreductase superfamily protein isoform 1 [Theobroma cacao] |
| 8 |
Hb_159305_010 |
0.1510233395 |
- |
- |
rpl2 [Jatropha curcas] |
| 9 |
Hb_012940_020 |
0.1512776949 |
- |
- |
zeaxanthin epoxidase, putative [Ricinus communis] |
| 10 |
Hb_000120_900 |
0.1520713431 |
- |
- |
catalase [Hevea brasiliensis] |
| 11 |
Hb_000667_140 |
0.1533811699 |
- |
- |
PREDICTED: uridine kinase-like protein 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_106552_030 |
0.1544210665 |
- |
- |
hypothetical protein POPTR_0006s23180g [Populus trichocarpa] |
| 13 |
Hb_000038_020 |
0.1544273287 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_189099_020 |
0.1544305301 |
- |
- |
NADH dehydrogenase subunit 9 (mitochondrion) [Hevea brasiliensis] |
| 15 |
Hb_034168_010 |
0.154884639 |
- |
- |
hypothetical protein [Azospirillum sp. CAG:260] |
| 16 |
Hb_003963_040 |
0.1551158492 |
- |
- |
ATP-dependent Clp protease proteolytic subunit [Hevea brasiliensis] |
| 17 |
Hb_185274_020 |
0.1571529178 |
- |
- |
Ycf2 (chloroplast) [Andrographis paniculata] |
| 18 |
Hb_000012_440 |
0.1580572824 |
- |
- |
PREDICTED: uncharacterized protein LOC104248335 [Nicotiana sylvestris] |
| 19 |
Hb_003032_090 |
0.1588615865 |
- |
- |
hypothetical protein POPTR_0018s06450g [Populus trichocarpa] |
| 20 |
Hb_156850_020 |
0.1596617429 |
- |
- |
ATP-dependent zinc metalloprotease FTSH 1, chloroplastic [Glycine soja] |