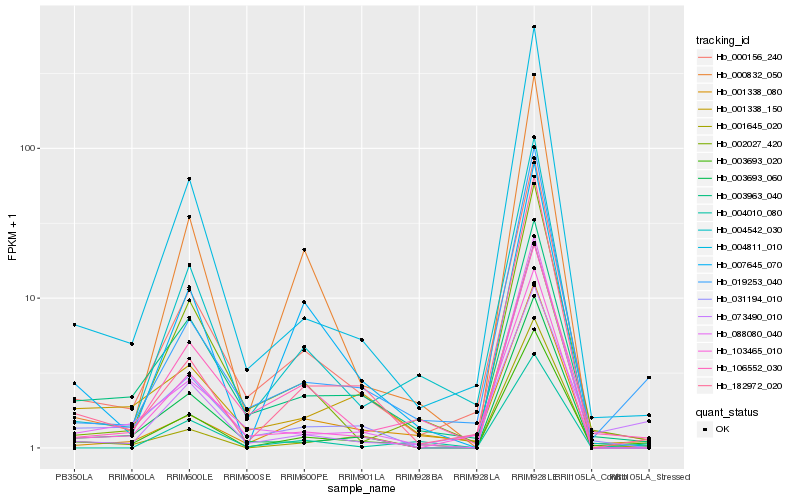
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003693_020 |
0.0 |
- |
- |
Ycf1, partial (chloroplast) [Clusia rosea] |
| 2 |
Hb_000156_240 |
0.1113915266 |
- |
- |
hypothetical protein MPER_07741 [Moniliophthora perniciosa FA553] |
| 3 |
Hb_031194_010 |
0.1233886544 |
- |
- |
NADH-plastoquinone oxidoreductase subunit 2 (chloroplast) [Arachis hypogaea] |
| 4 |
Hb_002027_420 |
0.1278474459 |
- |
- |
hypothetical protein POPTR_0020s00240g [Populus trichocarpa] |
| 5 |
Hb_003963_040 |
0.1295582754 |
- |
- |
ATP-dependent Clp protease proteolytic subunit [Hevea brasiliensis] |
| 6 |
Hb_019253_040 |
0.1306141667 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004542_030 |
0.1326248085 |
- |
- |
hypothetical protein AMTR_s00334p00014280 [Amborella trichopoda] |
| 8 |
Hb_001338_150 |
0.1344242129 |
- |
- |
hypothetical chloroplast RF1 [Dillenia indica] |
| 9 |
Hb_004811_010 |
0.1351084153 |
- |
- |
DNA-directed RNA polymerase subunit beta, partial [Medicago truncatula] |
| 10 |
Hb_106552_030 |
0.1380967767 |
- |
- |
hypothetical protein POPTR_0006s23180g [Populus trichocarpa] |
| 11 |
Hb_003693_060 |
0.1412248266 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit 6 [Medicago truncatula] |
| 12 |
Hb_073490_010 |
0.1433813876 |
- |
- |
NAD(P)-linked oxidoreductase superfamily protein isoform 1 [Theobroma cacao] |
| 13 |
Hb_001338_080 |
0.1465291123 |
- |
- |
NADH-plastoquinone oxidoreductase subunit 2 (chloroplast) [Tamarindus indica] |
| 14 |
Hb_007645_070 |
0.1503832053 |
- |
- |
photosystem II cytochrome b559 alpha subunit [Cuscuta obtusiflora] |
| 15 |
Hb_001645_020 |
0.1508803607 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit H [Medicago truncatula] |
| 16 |
Hb_004010_080 |
0.1521005558 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_182972_020 |
0.1545496245 |
- |
- |
cytochrome b6 [Gossypium barbadense] |
| 18 |
Hb_088080_040 |
0.1546531489 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit 2 A [Medicago truncatula] |
| 19 |
Hb_103465_010 |
0.1549181341 |
- |
- |
RNA polymerase beta subunit [Hevea brasiliensis] |
| 20 |
Hb_000832_050 |
0.1549581917 |
- |
- |
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit [Marila laxiflora] |