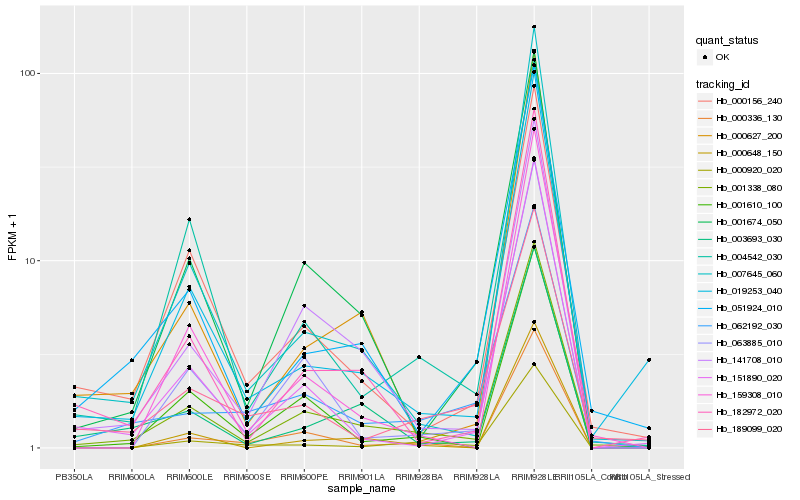
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001338_080 |
0.0 |
- |
- |
NADH-plastoquinone oxidoreductase subunit 2 (chloroplast) [Tamarindus indica] |
| 2 |
Hb_141708_010 |
0.0989921974 |
- |
- |
hypothetical protein EUTSA_v10011731mg [Eutrema salsugineum] |
| 3 |
Hb_001674_050 |
0.1020595959 |
- |
- |
cytochrome b6 (chloroplast) [Apios americana] |
| 4 |
Hb_051924_010 |
0.1035055299 |
- |
- |
BnaA01g34100D [Brassica napus] |
| 5 |
Hb_182972_020 |
0.1141203194 |
- |
- |
cytochrome b6 [Gossypium barbadense] |
| 6 |
Hb_019253_040 |
0.1186717922 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_007645_060 |
0.1204252971 |
- |
- |
cytochrome f [Hevea brasiliensis] |
| 8 |
Hb_000156_240 |
0.122642657 |
- |
- |
hypothetical protein MPER_07741 [Moniliophthora perniciosa FA553] |
| 9 |
Hb_001610_100 |
0.1257451 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_003693_030 |
0.1260190787 |
- |
- |
NADH dehydrogenase subunit 7 [Hevea brasiliensis] |
| 11 |
Hb_151890_020 |
0.1260216712 |
- |
- |
photosystem II cp47 protein, partial (chloroplast) [Pentaphragma sp. Duangjai 49] |
| 12 |
Hb_000627_200 |
0.1267313125 |
- |
- |
NADH dehydrogenase subunit I [Hevea brasiliensis] |
| 13 |
Hb_004542_030 |
0.1271399599 |
- |
- |
hypothetical protein AMTR_s00334p00014280 [Amborella trichopoda] |
| 14 |
Hb_063885_010 |
0.1294645091 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_062192_030 |
0.1304517851 |
- |
- |
ribosomal protein S1 (mitochondrion) [Hevea brasiliensis] |
| 16 |
Hb_000648_150 |
0.1329149754 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase V.1 [Camelina sativa] |
| 17 |
Hb_189099_020 |
0.1336179153 |
- |
- |
NADH dehydrogenase subunit 9 (mitochondrion) [Hevea brasiliensis] |
| 18 |
Hb_000920_020 |
0.1346147256 |
- |
- |
OSJNBa0004L19.22 [Oryza sativa Japonica Group] |
| 19 |
Hb_159308_010 |
0.1365827075 |
- |
- |
ribosomal protein S4 [Cercidiphyllum japonicum] |
| 20 |
Hb_000336_130 |
0.1366802793 |
- |
- |
Triacylglycerol lipase 2 precursor, putative [Ricinus communis] |