| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
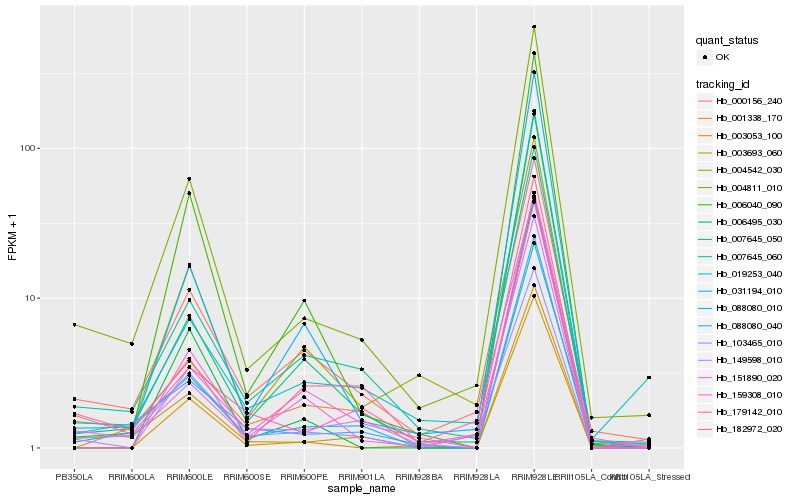
| 1 |
Hb_004811_010 |
0.0 |
- |
- |
DNA-directed RNA polymerase subunit beta, partial [Medicago truncatula] |
| 2 |
Hb_007645_060 |
0.0837830559 |
- |
- |
cytochrome f [Hevea brasiliensis] |
| 3 |
Hb_088080_040 |
0.0856734101 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit 2 A [Medicago truncatula] |
| 4 |
Hb_031194_010 |
0.0908536112 |
- |
- |
NADH-plastoquinone oxidoreductase subunit 2 (chloroplast) [Arachis hypogaea] |
| 5 |
Hb_006040_090 |
0.0961951675 |
- |
- |
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit [Hevea brasiliensis] |
| 6 |
Hb_149598_010 |
0.0973972802 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit 6 [Medicago truncatula] |
| 7 |
Hb_151890_020 |
0.1029708714 |
- |
- |
photosystem II cp47 protein, partial (chloroplast) [Pentaphragma sp. Duangjai 49] |
| 8 |
Hb_103465_010 |
0.1041491427 |
- |
- |
RNA polymerase beta subunit [Hevea brasiliensis] |
| 9 |
Hb_000156_240 |
0.1071979996 |
- |
- |
hypothetical protein MPER_07741 [Moniliophthora perniciosa FA553] |
| 10 |
Hb_003693_060 |
0.1072565442 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit 6 [Medicago truncatula] |
| 11 |
Hb_179142_010 |
0.108421172 |
- |
- |
NADH dehydrogenase ND 2 subunit [Nicotiana tomentosiformis] |
| 12 |
Hb_003053_100 |
0.1086550478 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 13 |
Hb_007645_050 |
0.1086763559 |
- |
- |
acetyl-CoA carboxylase beta subunit [Hevea brasiliensis] |
| 14 |
Hb_088080_010 |
0.1091806804 |
- |
- |
photosystem I P700 chlorophyll a apoprotein A1 [Matelea biflora] |
| 15 |
Hb_004542_030 |
0.1099493537 |
- |
- |
hypothetical protein AMTR_s00334p00014280 [Amborella trichopoda] |
| 16 |
Hb_006495_030 |
0.1105674274 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Vitis vinifera] |
| 17 |
Hb_001338_170 |
0.1106574857 |
- |
- |
NADH dehydrogenase subunit 4 [Hevea brasiliensis] |
| 18 |
Hb_182972_020 |
0.1115161505 |
- |
- |
cytochrome b6 [Gossypium barbadense] |
| 19 |
Hb_019253_040 |
0.1116595837 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_159308_010 |
0.1154585596 |
- |
- |
ribosomal protein S4 [Cercidiphyllum japonicum] |