| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
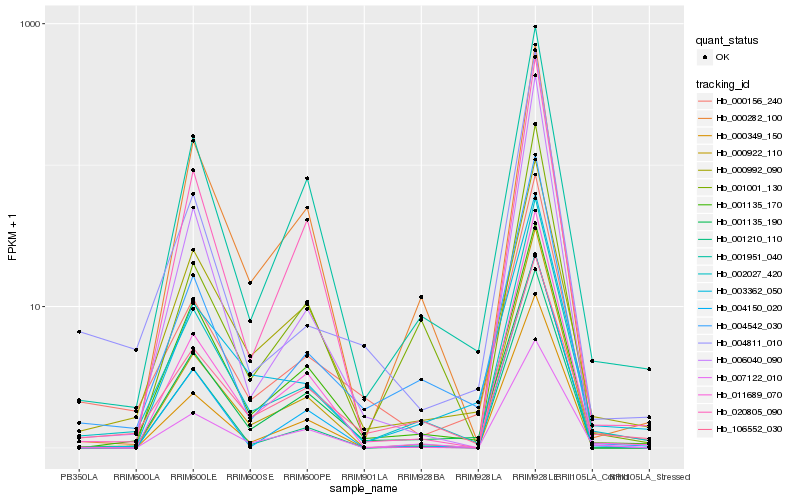
Hb_002027_420 |
0.0 |
- |
- |
hypothetical protein POPTR_0020s00240g [Populus trichocarpa] |
| 2 |
Hb_004542_030 |
0.0841764591 |
- |
- |
hypothetical protein AMTR_s00334p00014280 [Amborella trichopoda] |
| 3 |
Hb_001951_040 |
0.0994127006 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_007122_010 |
0.1009116759 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 5 |
Hb_000156_240 |
0.1013798267 |
- |
- |
hypothetical protein MPER_07741 [Moniliophthora perniciosa FA553] |
| 6 |
Hb_003362_050 |
0.1054439341 |
- |
- |
PREDICTED: probable sucrose-phosphate synthase 3 [Jatropha curcas] |
| 7 |
Hb_001135_190 |
0.107780579 |
transcription factor |
TF Family: Orphans |
Pseudo response regulator isoform 11 [Theobroma cacao] |
| 8 |
Hb_011689_070 |
0.1086694641 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000282_100 |
0.1109114228 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 10 |
Hb_000349_150 |
0.1112085646 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 11 |
Hb_000922_110 |
0.1122063759 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 12 |
Hb_006040_090 |
0.1129618799 |
- |
- |
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit [Hevea brasiliensis] |
| 13 |
Hb_000992_090 |
0.1150981881 |
transcription factor |
TF Family: Orphans |
CIL, putative [Ricinus communis] |
| 14 |
Hb_106552_030 |
0.1155011501 |
- |
- |
hypothetical protein POPTR_0006s23180g [Populus trichocarpa] |
| 15 |
Hb_001210_110 |
0.1160511837 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1 [Jatropha curcas] |
| 16 |
Hb_004811_010 |
0.1169905765 |
- |
- |
DNA-directed RNA polymerase subunit beta, partial [Medicago truncatula] |
| 17 |
Hb_001135_170 |
0.1182953437 |
transcription factor |
TF Family: Orphans |
Two-component response regulator ARR2, putative [Ricinus communis] |
| 18 |
Hb_020805_090 |
0.119830579 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_001001_130 |
0.1203617557 |
- |
- |
hypothetical protein POPTR_0002s12860g [Populus trichocarpa] |
| 20 |
Hb_004150_020 |
0.1203869809 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |