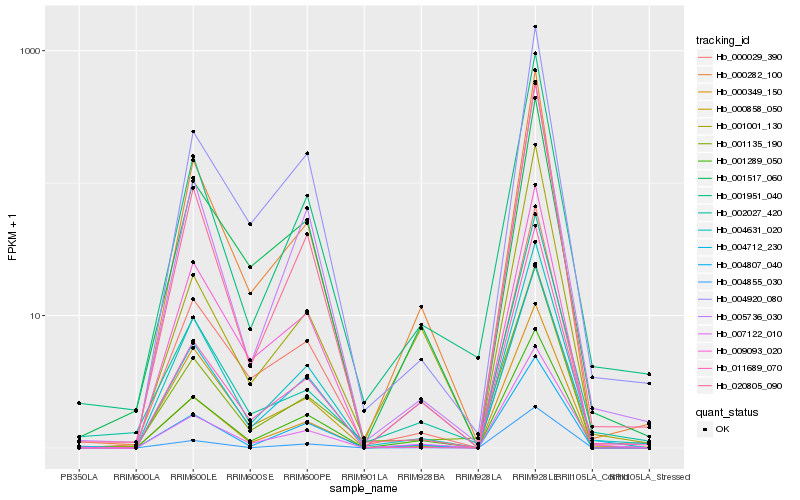
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007122_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 2 |
Hb_000282_100 |
0.0813253826 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 3 |
Hb_001951_040 |
0.0817575491 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_004712_230 |
0.0883841799 |
- |
- |
PREDICTED: retrotransposon-like protein 1 [Jatropha curcas] |
| 5 |
Hb_001289_050 |
0.0934887182 |
- |
- |
hypothetical protein Poptr_cp075 [Populus trichocarpa] |
| 6 |
Hb_000029_390 |
0.0944234719 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004631_020 |
0.0944749791 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 8 |
Hb_004855_030 |
0.0968119865 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 9 |
Hb_004920_080 |
0.0981240621 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_002027_420 |
0.1009116759 |
- |
- |
hypothetical protein POPTR_0020s00240g [Populus trichocarpa] |
| 11 |
Hb_005736_030 |
0.1018256095 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 12 |
Hb_000349_150 |
0.1022866257 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 13 |
Hb_020805_090 |
0.1035748203 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_011689_070 |
0.106295338 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000858_050 |
0.1078859041 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_001001_130 |
0.1091710907 |
- |
- |
hypothetical protein POPTR_0002s12860g [Populus trichocarpa] |
| 17 |
Hb_001135_190 |
0.1111281842 |
transcription factor |
TF Family: Orphans |
Pseudo response regulator isoform 11 [Theobroma cacao] |
| 18 |
Hb_009093_020 |
0.1114246691 |
- |
- |
PREDICTED: uncharacterized protein LOC105642100 [Jatropha curcas] |
| 19 |
Hb_004807_040 |
0.1121810382 |
- |
- |
PREDICTED: uncharacterized protein LOC104420559 [Eucalyptus grandis] |
| 20 |
Hb_001517_060 |
0.1122479131 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |