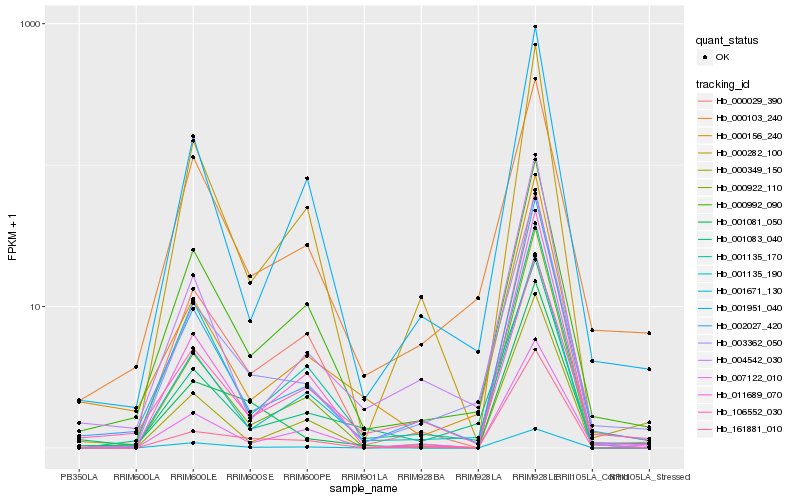
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003362_050 |
0.0 |
- |
- |
PREDICTED: probable sucrose-phosphate synthase 3 [Jatropha curcas] |
| 2 |
Hb_002027_420 |
0.1054439341 |
- |
- |
hypothetical protein POPTR_0020s00240g [Populus trichocarpa] |
| 3 |
Hb_001135_190 |
0.1166942365 |
transcription factor |
TF Family: Orphans |
Pseudo response regulator isoform 11 [Theobroma cacao] |
| 4 |
Hb_000992_090 |
0.1192130494 |
transcription factor |
TF Family: Orphans |
CIL, putative [Ricinus communis] |
| 5 |
Hb_000103_240 |
0.120055366 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 1 [Jatropha curcas] |
| 6 |
Hb_001083_040 |
0.1268886182 |
- |
- |
hypothetical protein EUGRSUZ_E00717 [Eucalyptus grandis] |
| 7 |
Hb_000029_390 |
0.1316076874 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001951_040 |
0.1327160131 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_004542_030 |
0.1331526286 |
- |
- |
hypothetical protein AMTR_s00334p00014280 [Amborella trichopoda] |
| 10 |
Hb_001135_170 |
0.1361095293 |
transcription factor |
TF Family: Orphans |
Two-component response regulator ARR2, putative [Ricinus communis] |
| 11 |
Hb_011689_070 |
0.1369444614 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000156_240 |
0.1382062664 |
- |
- |
hypothetical protein MPER_07741 [Moniliophthora perniciosa FA553] |
| 13 |
Hb_000282_100 |
0.138234016 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 14 |
Hb_007122_010 |
0.1385064383 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 15 |
Hb_161881_010 |
0.1424654659 |
- |
- |
laccase, putative [Ricinus communis] |
| 16 |
Hb_000922_110 |
0.1433128632 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 17 |
Hb_106552_030 |
0.1455801291 |
- |
- |
hypothetical protein POPTR_0006s23180g [Populus trichocarpa] |
| 18 |
Hb_001081_050 |
0.14691589 |
- |
- |
hypothetical protein POPTR_0001s07860g [Populus trichocarpa] |
| 19 |
Hb_001671_130 |
0.1481492223 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 20 |
Hb_000349_150 |
0.1490936514 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |